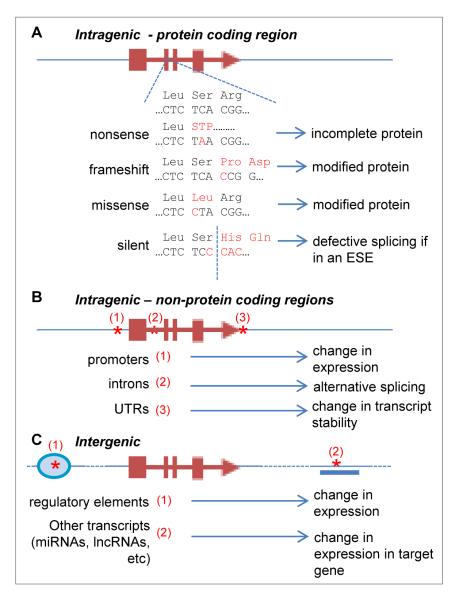
Fig. 2.
Potential mechanisms through which SNP variation can influence a target gene. Examples of how single-nucleotide variation can influence target genes. A. In protein-coding regions, SNPs can lead to changes in protein sequence leading to truncated or modified proteins with defective function or stability. Silent variation (i.e. nucleotide variation codes for the same amino acid residue) may disrupt exonic splicing enhancers (ESEs) leading to illegitimate or inefficient splicing. B. SNPs in intragenic (but non-protein-coding) regions can modify the activity of promoters, disrupt or create splicing acceptor and donor sites, and modify transcript instability in untranslated regions (UTRs), for example by influencing polyadenylation. C. Most GWAS hits are found in intergenic regions. SNPs in these regions may affect target genes by modifying regulatory sequences such as enhancers or insulator elements. SNPs may also modify other transcripts such as microRNAs (miRNAs) or long non-coding RNAs (lncRNAs) that may regulate other target genes.