Figure 1.
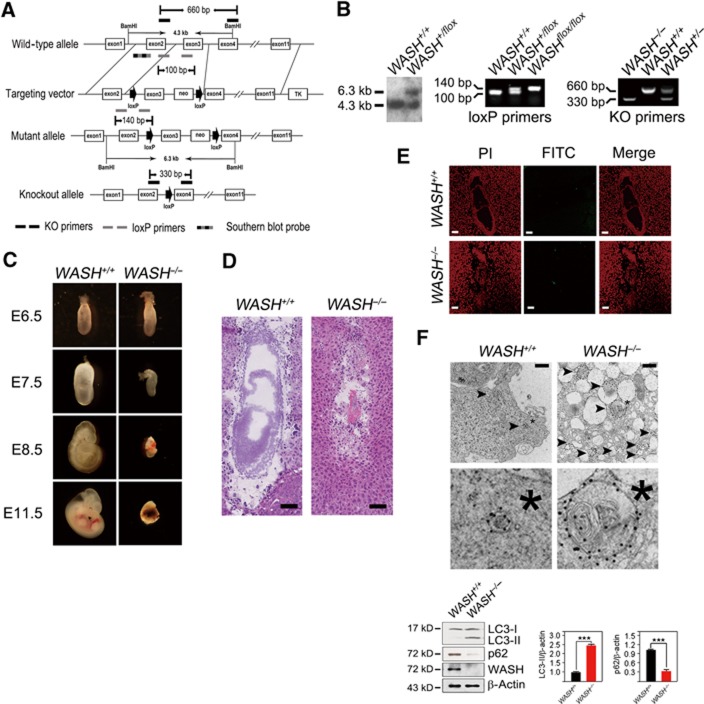
WASH deficiency causes embryonic lethality and extensive autophagy. (A) Strategy to generate WASH-knockout (WASH−/−) mice. The coding exons of mWASH gene were shown as white boxes. The target vector with exon3 of mWASH gene was flanked with two loxP sites (arrow) and one neomycin resistance gene. The primers used for genotyping were KO primers for deficient genotypes, loxP primers for loxP site identification, and Southern blot probe for floxed mWASH gene. KO, knockout, m, mouse. (B) Detection of WASH gene targeting. The floxed WASH gene by gene targeting was analysed by Southern blotting (left panel) and PCR (middle panel). The genotypes of the offspring were analysed by PCR (right panel). (C) WASH deficiency is embryonic lethal. E11.5 of WASH deficiency showed a shrinked decidua. (D) Histological analysis of E7.5 WASH+/+ and WASH−/− embryos. Scale bar, 100 μm. (E) WASH-deficient embryos do not undergo apoptosis. TUNEL assay was performed for apoptosis detection. PI, propidium iodide. FITC, fluoresceinisothiocyanate. Scale bar, 100 μm. (F) E7.5 embryos of WASH−/− mice present extensive autophagy. Ultrastructure of isolated E7.5 embryos was stained with antibody against LC3 and visualized by immuno-electron microscopy. Black arrowhead indicates autophagosome. *, Cellular reference. Scale bar, 500 nm (upper panel). WASH+/+ or WASH−/− embryos were isolated, and LC3 conversion and p62 level were analysed by immunoblotting (lower panel). Bands were quantified and analysed by Image J and shown as means±s.d. ***P<0.001. Above experiments were repeated for three independent times with similar results.
Source data for this figure is available on the online supplementary information page.