Figure 1.
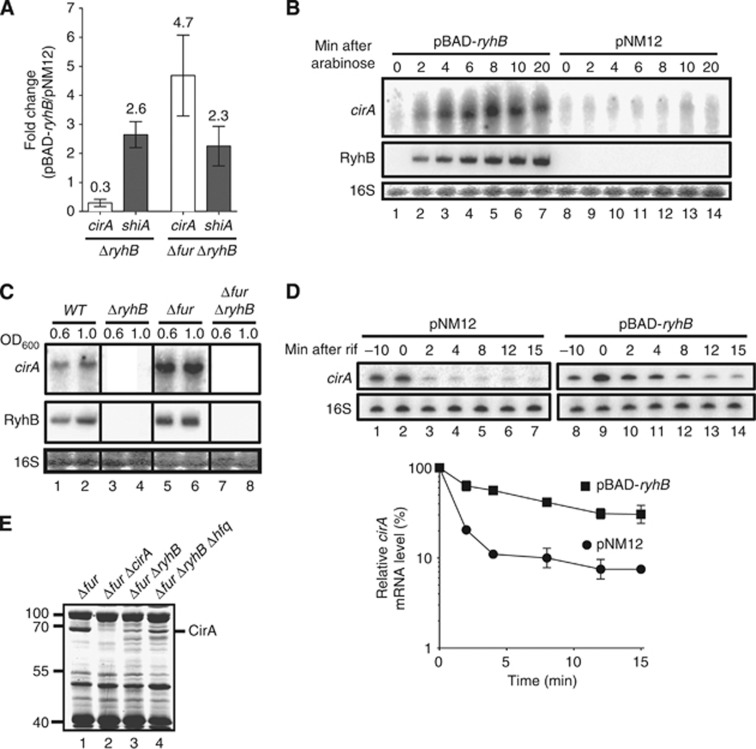
RyhB expression is essential for CirA production. (A) Quantitative RT-PCR (qRT-PCR) analysis of RyhB effect on cirA and shiA mRNA levels. Strains EM1455 (ΔryhB) and EM1493 (Δfur ΔryhB), each carrying either pBAD-ryhB or control plasmid pNM12, were grown in LB medium. Arabinose was added (0.1%, total concentration) to an OD600 of 0.5, followed by total RNA extracted after 20 min. Primers complementary to cirA and shiA open reading frames (ORFs) were used for qRT–PCR assays. Mean and standard deviation (s.d.) values of triplicate samples are shown. (B) Northern blot analysis of RyhB effect on cirA mRNA levels. Strain EM1493 (Δfur ΔryhB) carrying pBAD-ryhB or control plasmid pNM12 was grown in LB medium. Arabinose was added (0.1%, total concentration) to an OD600 of 0.5 and total RNA was extracted at the indicated time points. A probe complementary to cirA ORF was used. (C) Northern blot analysis of RyhB and Fur effects on cirA mRNA levels in cells grown under iron-poor conditions. Strains EM1055 (WT), EM1238 (ΔryhB), KP392 (Δfur) and KP393 (Δfur ΔryhB) were grown in M63 iron-free glucose minimal medium and total RNA was extracted at the indicated values of OD600. A probe complementary to cirA ORF was used. (D) Primer extension analysis of RyhB effect on cirA mRNA stability. Strain EM1493 (Δfur ΔryhB) carrying pBAD-ryhB or control plasmid pNM12 was grown in LB medium and arabinose was added (0.1%, total concentration) to an OD600 of 0.5 (−10 time point). Rifampicin (250 μg/ml, final concentration) was added 10 min after the addition of arabinose (0 time point) and total RNA was extracted at the indicated time points. Primers complementary to cirA (EM1408) and 16S rRNA (EM345) were used to perform primer extension reactions on total RNA samples. Primer extension signals of two independent experiments were quantified by densitometry and normalized to time zero (0) for both pNM12 and pBAD-ryhB. Mean and s.d. values of duplicate experiments are shown. (E) Coomassie-stained SDS gel of outer membrane proteins (OMPs) extracts from KP392 (Δfur), HS437 (Δfur ΔcirA), KP393 (Δfur ΔryhB) and MPC253 (Δfur ΔryhB Δhfq) cells grown in LB medium at an OD600 of 1.0. Molecular sizes of co-migrating proteins (L) are indicated at the left in kDa.
Source data for this figure is available on the online supplementary information page.