Figure 3.
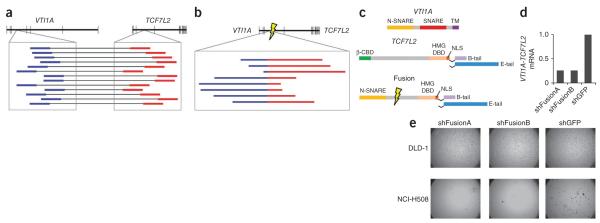
Recurrent gene fusion between VTI1A and TCF7L2. (a) The upper schematic depicts the positions of exons (vertical lines) within VTI1A and TCF7L2, which reside adjacent to each other on chromosome 10. The blowup displays the locations of discordant paired-end reads found in tumor CRC-9 for which one read (labeled in blue) is in an intron of VTI1A and the other read (labeled in red) is in an intron of TCF7L2. (b) The upper schematic depicts the structure of the predicted fusion transcript generated by the fusion. The presence of the exact reads spanning the fusion of the two introns (marked by lightning bolt) is depicted in the inset with regions of the reads corresponding to original VTI1A intron in blue and those of TCF7L2 in red. (c) The protein domain structure of native VTI1A and TCF4-TCF7L2, including the two alternate C-terminal tails of TCF4, are shown. Below are the structures of the fusion protein encoded by the fusion of exon 3 of VTI1A to exon 4 of TCF7L2 identified in CRC-9. Two variants of the fusion are shown as data from the NCI-H508 cell line and reveal that variants encoding both the full length (E-tail) and shorter (B-tail) C termini are both expressed (data not shown). (d) Measurement of the relative expression of the VTI1A-TCF7L2 mRNA in NCI-H508 cells infected with one of two short hairpin RNA constructs targeting the fusion gene relative to expression in a cell infected with control vectors targeting GFP. (e) Anchorage-independent growth of the NCIH508 cell line, which expresses VTI1A-TCF7L2, and negative control DLD-1 colorectal adenocarcinoma cells following RNA-interference–mediated knockdown of VTI1A-TCF7L2 compared to control knockdown targeting GFP.
