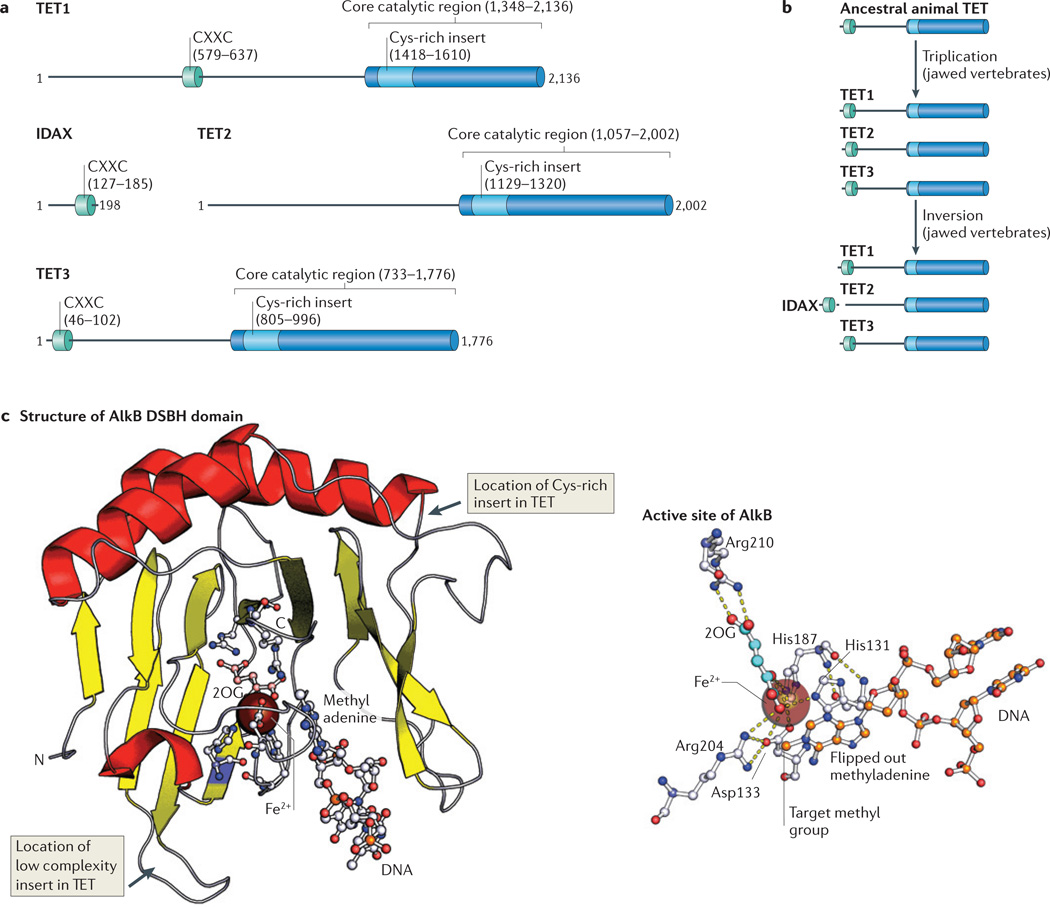
Figure 2. Known protein domains of TET family members.
a | Ten-eleven translocation (TET) proteins contain a DNA-binding CXXC domain towards the ami no terminus and a carboxy-terminal catalytic core region that includes a Cys-rich insert and a larger double-stranded β-helix (DSBH) domain. The number of amino acids is indicated, and the numbering corresponds to the human proteins. b | Evolutionary changes in the domain structure of TET proteins. A gene triplication event that occurred in jawed vertebrates resulted in the generation of three TET family members. A chromosomal inversion then detached the catalytic domain of TET2 from its CXXC domain, which became a separate gene (which encodes IDAX (inhibition of the Dvl and axin complex)). c | A cartoon representation of AlkB (Protein Databank (PDB) identifier: 2FD8), a protein that belongs to the same superfamily as the TET proteins and shares a common fold with them. AlkB is shown as a complex with its substrate methyladenine and its cofactor 2-oxoglutarate (2OG). In the 2OG structure, carbon atoms are shown pink, in the rest of the structure carbons are shown in grey. Nitrogens are shown in blue, oxygens in red and phosphates in orange (left panel). A stripped-down view of the active site of AlkB in complex with its substrate methlyadenine. Note the series of interactions, including pi–pi stacking interactions, between the His residues and the target base, and cation–pi interaction with the active site metal. Such interactions are likely to be retained in the TET– J-binding protein (JBP) family. In the 2OG structure, carbon atoms are shown in cyan and oxygens in red. In the protein structure, carbons are shown in grey, nitrogens in blue and oxygens in red. In methyladenine, carbons are orange and nitrogens are blue. Dashed yellow lines represent hydrogen bonds (right panel).