Abstract
Salmonella enterica serovar Enteritidis is one of the main pathogens responsible for foodborne illness in Brazil. Probiotic bacteria can play a role in defense and recovery from enteropathogenic infections. In this study, the ability of Lactobacillus acidophilus LA10 to colonise and exert antagonistic effects in the gastrointestinal tract was tested before and during experimental infection in conventional mice contaminated with S. Enteritidis (SE86). A dose of 0.1 mL containing 108 viable cells of SE86 and L. acidophilus LA10 was orally administered by gavage to mice. The experiment was divided into groups. As a negative control, Group 1 was administered only sterile saline solution. As a positive control, Group 2 was administered only SE86. Group 3 was first administered SE86, and after 10 days, treated with L. acidophilus LA10. Group 4 was first administered L. acidophilus LA10, and after 10 days, challenged with SE86. The results demonstrated that a significant number of SE86 cells were able to colonize the gastrointestinal tract of mice, specifically in the colon and ileum. L. acidophilus LA10 demonstrated an antagonistic effect against SE86, with better results observed for Group 3 over Group 4. Thus, L. acidophilus LA10 shows potential antagonistic effects against S. Enteritidis SE86, especially if administered after infection.
Keywords: S Enteritidis SE86, probiotics, mice, Lactobacillus acidophilus LA10
Introduction
Salmonella spp. has long been recognised as a common cause of foodborne gastroenteritis in humans (Centers for Disease Control and Prevention, 2009) and responsible for significant economic losses in the food industry (Humphrey, 2004). Although Salmonella gastroenteritis may be caused by any of more than 2,500 serotypes (Kangas et al., 2007), S. Enteritidis remains one of the main causes of foodborne illness, and is considered one of the most important pandemic zoonosis produced under natural conditions (Araya et al., 2010; Matheson et al., 2010). It has been reported that a well characterised pathogenic S. Enteritidis strain (SE86) was involved in many foodborne outbreaks in the State of Rio Grande do Sul (RS), the southernmost state in Brazil (Geimba et al., 2004; Moura et al., 2001; Oelschlaeger, 2010). In addition, SE86 has been reported to undergo acid adaptation, as characterised by: increased acid and thermal resistance (Araya et al., 2010) higher survival rates in simulated gastric fluid (Bernardeau et al., 2008), and better intestinal colonisation in mice (Borowsky et al. 2007; Malheiros et al., 2009; Perez et al., 2010).
The reduction of infections caused by foodborne pathogens such as S. Enteritidis is of great importance for public health. Treatment of salmonellosis is carried out mainly with fluid replacement, and the antibiotics are only recommended for extra-intestinal infections and exceptional cases. The possibility of using probiotic bacteria for protection against enteropathogenic infections has been considered (Gibson et al., 2005; Roselli et al., 2006). Probiotics are defined as live microorganisms that, when ingested in appropriate amounts, confer a health benefit to the host (FAO/WHO, 2001). Probiotic bacteria can play a role in defense and recovery from enteropathogenic infections (Candela et al., 2008; Servin, 2004), especially by protecting the host from enteropathogen colonization, and by modulating the host immune response (Resta-Lenert and Barrett, 2003).
The most widely used probiotic bacteria, and more relevant to the prevention of tissue infection by enteropathogens are Bifidobacterium and Lactobacillus (Candela et al., 2008; Fooks and Gibson, 2002; Guarner and Malgelada, 2003). Among the Lactobacillus species, L. acidophilus, L. plantarum, L. bulgaricus, L. casei, and L. rhamnosus have been frequently used. These Gram positive bacteria are natural inhabitants of the gastrointestinal tract, which they colonize by adhering to the intestinal epithelium, where they produce lactic acid and effectively act as probiotics (Bernardeau et al., 2008). L. acidophilus has been reported to be capable of stimulating the defense mechanisms of the immune system by colonizing the gastrointestinal tract and preventing the adhesion of many enterotoxigenic and enteroinvasive bacteria (Moura et al., 2001).
Based on these findings, the goal of this study was to evaluate the ability of a probiotic L. acidophilus LA10 to exert antagonistic effects against S. Enteritidis SE86 in mice.
Materials and Methods
Animal handling and experimental protocol
Conventional male Swiss mice, 21–23 days of age, were used in this work. The animals were individually housed, supplied with potable water and commercial animal feed ad libitum, at 22 °C ± 2 °C, with 65% at 70% humidity and alternate 12 h periods of light and dark. All experimental procedures were carried out according to standards set forth by the Ethics and Research Council of the Universidade do Oeste de Santa Catarina, protocol number 017/2008.
Bacterial strains and culture conditions
S. Enteritidis SE86 was isolated from a cabbage involved in a foodborne outbreak occurred in 1999 in Rio Grande do Sul (RS) State, in Brazil. This strain shows the same genotypic profile as more than 90% of the S. Enteritidis involved in foodborne salmonellosis in RS State during the period between 1999 and 2002 (Geimba et al., 2004; Oliveira et al., 2007). Before the experiments, the SE86 was stored at −18 °C in 50% (v/v) glycerol. Working cultures were kept at 4 °C on BHI agar plates (Merck, Darmstadt, Germany), and subcultured in tubes containing BHI (Merck, Darmstadt, Germany) broth at 37 °C for 24 h.
Capsules of liophylized L. acidophilus LA10 were acquired in a pharmacy located in São Miguel do Oeste, State of Santa Catarina, Brazil. The capsule contents were diluted in 1 mL of sterile distilled water resulting in 108 cells/mL.
Treatments and experimental infection
A single dose of 0.1 mL containing 108 viable cells of SE86 was orally administered to each mice. A dose of 0.1 mL containing 2 × 108 cells/mL of L. acidophilus LA10 was administered by gavage (Bambirra et al., 2007).
As a negative control, Group 1 mice were only administered sterile saline solution. As a positive control, Group 2 mice were administered only S. Enteritidis SE86. Mice from both groups were sacrificed after 10 days.
Experimental groups were divided into two (Group 3 and Group 4). Group 3 mice were first challenged with S. Enteritidis SE86, and after 10 days, were treated with L. acidophilus LA10. Group 4 mice were first administered with L. acidophilus LA10, and after 10 days, were challenged with S. Enteritidis SE86. Both groups were sacrificed 10 days after administration of the last microorganism.
Microbial counts
Fresh mice feces (1 g) were collected on the tenth day after administration of the microorganisms. Portions of the gastrointestinal tract (0.1 g of ileum and colon) were sampled at the tenth day after mice were sacrificed by cervical dislocation. The ileum and colon portions were homogenized into tubes containing 9.99 mL of buffered saline peptone water, while feces were homogenized in 9 mL of the same solution. Samples were homogenized for 2 min using an automatic mixer. Decimal dilutions were prepared and L. acidophilus LA10 cell counts were performed by spreading 1 mL of diluted feces (0.1 mL) or intestinal portions (0.01 mL) on plates containing Agar-Man-Rogosa Sharpe (MRS) (Merck, Darmstadt, Germany) and the plates were incubated at 36 °C for 48–72 h in a microaerophilic incubator (Moura et al., 2001; Silva, Junqueira and Silveira 1997). Typical colonies were confirmed using biochemical tests (Macfaddin, 2000).
Microbiological quantification of SE86 was carried out using the most probable number (MPN) method (Borowsky et al., 2007).
Statistical analysis
Statistical analysis was performed using the Chi-square and Pearson correlation coefficient to assess population levels. The level of significance was set at p ≤ 0.05 using the statistical package SPSS, Version 12 for Windows.
Results and Discussion
Our results confirmed that mice from Group 1 (negative control) did not contain SE86 in the gastrointestinal tract or in feces. In contrast, in Group 2 mice (positive control) challenged with SE86, the colon was clearly colonized, with counts between < 30/g and > 11000/g (arithmetic mean, 3760 MPN/g). Feces from these Group 2 mice displayed counts varying from 9.2 to > 1100/g (arithmetic mean, 593 MPN/g), and ileum counts varying from < 30/g to 230/g (arithmetic mean, 78 MPN/g). Similar results were reported by Perez et al. (2010), who found that SE86 was able to infect the gastrointestinal tract of rats and spread into the feces.
According to Llana et al. (2009), salmonellosis in the rat has many similarities with the disease in humans. The study of Lahiri et al. (2010) showed that, immediately after infection, Salmonella were found preferentially associated with Peyer’s patches in the terminal ileum, which is thought to be the main site of colonization/invasion. Thus, rats may be useful to study mechanisms of infection by these pathogenic bacteria (Rodenburg et al., 2007b). Rodenburg et al. (2007a) reported that Salmonella tend to remain in the gastrointestinal tract during all stages of infection, and that a relatively high number of cells persist in the intestine after contamination with the pathogenic bacteria. The same researchers Rodenburg et al. (2007a) suggested that S. Enteritidis is able to translocate to the small intestine, and induce gene expression changes in the ileal mucosa and Peyer’s patches. However, the effects of Salmonella on colonic gene expression in vivo are largely unknown.
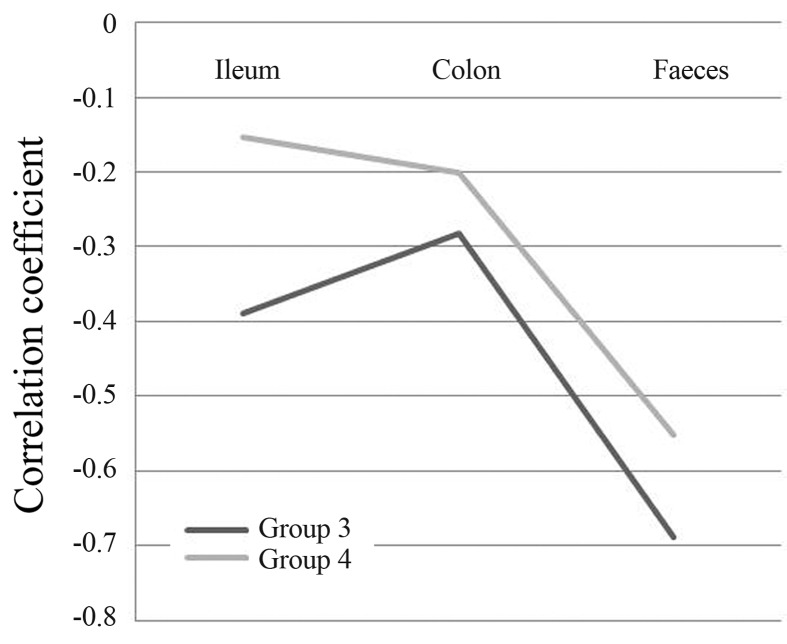
In Group 3 (which was first challenged with S. Enteritidis SE86 and after 10 days treated with L. acidophilus LA10 we observed that the mice showed no SE86 in their intestinal portions (either the ileum or colon). In this group, the mice eliminated the pathogenic bacteria in their feces (Table 1). The correlation between counts of L. acidophillus LA10 and S. Enteritidis SE86 was inversely proportional (Figure 1), with a medium high negative correlation for feces (−0.6831), and medium negative correlation in the colon (−0.2822) and ileum (−0.3893).
Table 1.
Relationship between counts of Salmonella Enteritidis SE86 and Lactobacillus acidophilus LA10 in portions of the intestine and feces of mice from the group initially challenged with Salmonella Enteritidis SE86 and treated after 10 days with Lactobacillus acidophilus LA10.
| Animals | Salmonella Enteritidis SE86 (MPN/g) | Lactobacillus acidophilus LA10 (CFU/g) | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Ileum | Colon | Feces | Ileum | Colon | Feces | |
| Mice #1 | < 30* | < 30 | 7.4 | 3.76 × 106 | 1.0 × 102 | 8.83 × 107 |
| Mice #2 | < 30 | < 30 | 43 | 7.28 × 105 | 7.65 × 103 | 3.05 × 107 |
| Mice #3 | < 30 | < 30 | 23 | 1.99 × 106 | 2.75 × 108 | 2.15 × 107 |
| Mice #4 | < 30 | < 30 | < 3.0 | 4.21 × 106 | 1.15 × 104 | 5.4 × 107 |
| Mice #5 | 430 | 930 | < 3.0 | 1.43 × 106 | 2.8 × 107 | 1.85 × 107 |
| Mice #6 | < 30 | < 30 | < 3.0 | 2.95 × 106 | 1.34 × 108 | 4.1 × 107 |
For statistical analysis purposes it was considered the maximum number of 11000 for counts that have the sign > (greater than) and minimum of 3.0 for counts that have the sign < (less than) to feces and portions of the intestine, respectively.
Figure 1.
Correlation between the MPN/g of Salmonella Enteritidis SE86 and Lactobacillus acidophilus LA-10 counts (CFU/g) in portions of the intestines and feces of mice in the two evaluated groups.
In Group 4 (which was first treated with L. acidophilus LA10 and after 10 days challenged with S. Enteritidis SE86), colonization by pathogenic bacteria was higher in the colon, followed by the ileum and feces (Table 2). The correlation between counts of L. acidophilus LA10 and S. Enteritidis SE86 was again inversely proportional (Figure 1) as in Group 3, with a medium high negative correlation in the feces (−0.5524), and a medium negative correlation in the colon (−0.2011) and ileum (−0.1527).
Table 2.
Relationship between counts of Salmonella Enteritidis SE86 and Lactobacillus acidophilus LA10 in the gastrointestinal tract and feces of mice from the group treated initially with Lactobacillus acidophilus LA10 and after 10 days challenged with Salmonella Enteritidis SE86.
| Animals | Salmonella Enteritidis SE86 (MPN/g) | Lactobacillus acidophilus LA10 (CFU/g) | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Ileum | Colon | Feces | Ileum | Colon | Feces | |
| Mice #1 | 92 | > 11000 | 23 | 1.38 × 107 | 4.75 × 104 | 1.14 × 108 |
| Mice #2 | < 30 | 61 | < 3.0 | 2.51 × 107 | 8.45 × 104 | 2.93 × 107 |
| Mice #3 | 2100 | < 30 | 460 | 4.55 × 106 | 6.75 × 106 | 1.16 × 107 |
| Mice #4 | < 30 | < 30 | < 3.0 | 1.92 × 106 | 1.2 × 104 | 2.45 × 107 |
| Mice #5 | < 30 | < 30 | 23 | 3.44 × 105 | < 1.0 × 102 | 1.08 × 108 |
| Mice #6 | < 30 | 200 | 240 | 9.44 × 105 | < 1.0 × 102 | 1.9 × 107 |
For statistical analysis purposes it was considered the maximum number of 11000 for counts that have the sign > (greater than) and minimum of 3.0 for counts that have the sign < (less than) to feces and portions of the intestine, respectively.
Havelaar et al. (2001) reported that intestinal colonization by S. Enteritidis is concentrated in the distal ileum and caecum, and may be detected by fecal excretion. Lahiri et al. (2010) suggested that the terminal ileum is the primary site of Salmonella infection. However, Rodenburg et al. (2007a) showed that, in addition to the ileum, the colon mucosa is clearly a target for Salmonella infection, which could explain the increased colonization by S. Enteritidis SE86 found in the colon in this study (group 4). Also, Vender and Marignani (1983), found multiple ulcerations caused by Salmonella in the distal transverse colon and proximal descending colon in biopsies performed on humans with salmonellosis, highlighting the importance of studies investigating the infection process of this bacterium in the gastrointestinal tract, and specifically in the colon.
Comparing results obtained in Groups 3 and 4, we observed that after treatment with L. acidophilus LA10, the MPN of S. Enteritidis SE86 in the latter group was higher than in the former, although the counts of L. acidophilus LA10 remained in both groups between 106 and 107 cfu/g. These data were also confirmed statistically, and the correlation coefficient was higher in Group 3 in all intestinal portions evaluated, as well as in the feces (Figure 1).
The fact that the NMP/g of SE86 was lower in the experimental group first challenged with S. Enteritidis SE86 and after 10 days treated with L. acidophilus LA10 (Group 3) suggests that L. acidophilus LA10 is able to proliferate in the conditions of the gastrointestinal tract and colonize in high population levels in the gut ecosystem, demonstrating that this probiotic is an effective antagonist against S. Enteritidis SE86. The antagonistic activity of lactic acid bacteria against Salmonella infection has been studied elsewhere (Coconnier et al., 2000). Statistically, the best inverse correlation results were found in this group (Figure 1), emphasising that the administration of L. acidophilus LA10 after infection by S. Enteritidis SE86 showed better antagonist effects against this pathogenic bacteria (Tables 1 and 2).
The protection offered by L. acidophilus LA10 against pathogenic bacteria challenge is probably due to additional protection mechanisms provided by these intestinal microbiota and their properties as bio-therapeutic agents (Oelschlaeger, 2010). Similar results were reported by Moura et al. (2001) and Silva et al. (1999) using L. acidophilus and Bifidobacterium, respectively. These authors observed that probiotics produce antagonistic substances against S. Enteritidis. However, this protection is not due to a reduction of the pathogenic populations in the intestines, but instead this bio-therapeutic agent may act through other protective mechanisms such as immunomodulation. In addition, other properties, such as competition for adherence sites, may also explain the protective effects of Lactobacillus against enteropathogenic bacteria (Oelschlaeger, 2010).
Conclusions
In conclusion, L. acidophilus LA10 showed antagonistic effect against S. Enteritidis SE86, suggesting that this probiotic can be used as a therapeutic tool against salmonellosis. This is an interesting result since S. Enteritidis SE86 has been identified as responsible for salmonellosis outbreaks since 1999 in Rio Grande do Sul State, southern Brazil.
Furthermore, the administration of probiotics during infection by S. Enteritidis SE86 (treatment) seems to be more effective than administration before infection (prevention).
References
- Araya DV, Quiroz TS, Tobar HE, Lizana RL, Quezada CP, Santiviago CA, Riedel CA, Kalergis AM, Bueno SM. Deletion of a prophage-like element causes attenuation of Salmonella enterica serovar Enteritidis and promotes protective immunity. Vaccine. 2010;28:5458–5466. doi: 10.1016/j.vaccine.2010.05.073. [DOI] [PubMed] [Google Scholar]
- Bambirra FHS, Lima KGC, Franco BDGM, Cara DC, Nardi RMD, Barbosa FHF, Nicoli JR. Protective effect of Lactobacillus sakei 2a against experimental challenge with Listeria monocytogenes in gnotobiotic mice. Lett Appl Microbiol. 2007;45:663–667. doi: 10.1111/j.1472-765X.2007.02250.x. [DOI] [PubMed] [Google Scholar]
- Bernardeau M, Vernoux JP, Henri-Dubernet S, Guéguen M. Safety assessment of dairy microorganisms: The Lactobacillus genus. Int J Food Microbiol. 2008;126:278–285. doi: 10.1016/j.ijfoodmicro.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Borowsky LM, Schmidt V, Cardoso M. Estimation of most probable number of salmonella in minced pork samples. Braz J Microbiol. 2007;38:544–546. [Google Scholar]
- Candela M, Perna F, Carnevali P, Vitali B, Ciati R, Gionchetti P, Rizello F, Campieri M, Bridigi P. Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: Adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int J Food Microbiol. 2008;125:286–292. doi: 10.1016/j.ijfoodmicro.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Preliminary FoodNet dataon the incidence of infection with pathogens transmitted commonly through food-10 states, 2008. Morb Mortal Wkly Rep. 2009;58:333–337. [PubMed] [Google Scholar]
- Coconnier MH, Liévin V, Lorrot M, Servin AL. Antagonistic activity of Lactobacillus acidophilus LB against intracellular Salmonella enterica serovar Typhimurium infecting human enterocyte-like Caco-2/TC- 7 cells. Appl Environ Microbiol. 2000;66:1152–1157. doi: 10.1128/aem.66.3.1152-1157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food and Agriculture Organization of United Nations, World Health Organization. Evaluation of health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Report of a joint FAO/WHO expert consultation, Córdoba, Argentina. 2001 ftp://ftp.fao.org/es/esn/food/probioreport_en.pdf.
- Fooks LJ, Gibson GR. Probiotics as modulators of the gut flora. Br J Nutr. 2002;88:39–49. doi: 10.1079/BJN2002628. [DOI] [PubMed] [Google Scholar]
- Geimba MP, Tondo EC, Oliveira FA, Canal CW, Brandelli A. Serological characterization and prevalence of spvR genes in Salmonella isolated from foods involved in outbreaks in Brazil. J Food Prot. 2004;67:1229–1233. doi: 10.4315/0362-028x-67.6.1229. [DOI] [PubMed] [Google Scholar]
- Gibson GR, McCartney AL, Rastall RA. Prebiotics and resistance to gastrointestinal infections. Br J Nutr. 2005;93:31–34. doi: 10.1079/bjn20041343. [DOI] [PubMed] [Google Scholar]
- Guarner F, Malgelada JR. Gut flora in health and disease. Lancet. 2003;361:512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- Havelaar AH, Garssen J, Takumi K, Koedam MA, Dufrenne JB, Van Leusden FM, La Fonteyne L, Bousema JT, Vos JG. A rat model for dose-response relationships of Salmonella Enteritidis infection. J Appl Microbiol. 2001;91:442–452. doi: 10.1046/j.1365-2672.2001.01399.x. [DOI] [PubMed] [Google Scholar]
- Humphrey T. Salmonella stress responses and food safety. Science and Society. 2004;2:504–509. doi: 10.1038/nrmicro907. [DOI] [PubMed] [Google Scholar]
- Kangas S, Lyytikainen T, Peltola J, Ranta J, Maijala R. Costs of two alternative Salmonella control policies in Finnish broiler production. Acta Vet Scand. 2007;49:01–08. doi: 10.1186/1751-0147-49-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri A, Lahiri A, Iyer N, Das P, Chakravortty D. Visiting the cell biology of Salmonella infection. Microbes Infect. 2010;12:809–818. doi: 10.1016/j.micinf.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Llana MN, Sarnacki SH, Giacomodonato MN, Caccuri RL, Blanco GA, Cerquetti MC. Sublethal infection with Salmonella Enteritidis by the natural route induces intestinal and joint inflammation in mice. Microbes Infect. 2009;11:74–82. doi: 10.1016/j.micinf.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Macfaddin JF. Biochemical Tests for Identifications of Medical Bacteria. 3rd ed. Lippincott Williams & Wilkins; Philadelphia: 2000. p. 912. [Google Scholar]
- Malheiros PS, Brandelli A, Norena CPZ, Tondo EC. Acid and thermal resistance of a Salmonella Enteritidis strain involved in several foodborne outbreaks. J Food Safety. 2009;29:302–317. [Google Scholar]
- Matheson N, Kingsley R, Sturgess K, Aliyu S, Wain J, Dougan G. Ten years experience of Salmonella infections in Cambridge, UK. J Infect. 2010;60:21–25. doi: 10.1016/j.jinf.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Moura LN, Neumann E, Vieira LQ, Nicoli JR. Protection by Lactobacillus acidophilus UFV-H2B20 against experimental oral infection with Salmonella enterica subsp. enterica ser. Typhymurium in gnotobiotic and convencional mice. Braz J Microbiol. 2001;32:66–69. [Google Scholar]
- Oliveira FA, Frazzon APG, Brandelli A, Tondo EC. Use of PCR-Ribotyping, RAPD, and Antimicrobial Resistance for typing of Salmonella Enteritidis involved in foodborne outbreaks in Southern Brazil. J Infect Developing Countries. 2007;1:170–176. [Google Scholar]
- Oelschlaeger TA. Mechanisms of probiotic actions - A review. Int J Med Microbiol. 2010;300:57–62. doi: 10.1016/j.ijmm.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Pasquali G, Silva WP, Tondo EC. Clonal relationship among Salmonella enterica serovar Enteritidis involved in foodborne outbreaks in Sourthern Brazil. Food Control. 2009;20:606–610. [Google Scholar]
- Perez KJ, Ceccon RV, Malheiros PS, Jong EV, Tondo EC. Influence of acid adaptation on the survival of Salmonella Enteritidis and Salmonella Typhimurium in simulated gastric fluid and in intestine infection. J Food Safety. 2010;30:398–414. [Google Scholar]
- Resta-Lenert S, Barrett KE. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC) Int J Gastroenterol Hepatol. 2003;52:988–997. doi: 10.1136/gut.52.7.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenburg W, Keijer J, Kramer E, Roosing S, Vink C, Katan MB, Van der Meer R, Bovee-Oudenhoven MJ. Salmonella induces prominent gene expression in the rat colon. BMC Microbiol. 2007a;7:1–16. doi: 10.1186/1471-2180-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenburg W, Bovee-Oudenhoven IM, Kramer E, Van der Meer R, Keijer J. Gene expression response of the rat small intestine following oral Salmonella infection. Physiol Genomics. 2007b;30:123–133. doi: 10.1152/physiolgenomics.00190.2006. [DOI] [PubMed] [Google Scholar]
- Roselli M, Finamore A, Britti MS, Mengheri E. Probiotic bacteria Bifidobacterium animalis MB5 and Lactobacillus rhamnosus GG protect intestinal Caco-2 cells from the inflammation-associated response induced by enterotoxigenic Escherichia coli K88. Br J Nutr. 2006;95:1177–1184. doi: 10.1079/bjn20051681. [DOI] [PubMed] [Google Scholar]
- Servin AL. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. 2004;28:405–440. doi: 10.1016/j.femsre.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Silva AM, Bambirra EA, Oliveira AL, Souza PP, Gomes DA, Vieira EC, Nicoli JR. Protective effect of bifidus milk on the experimental infection with Salmonella enteritidis subsp. typhimurium in conventional and gnotobiotic mice. J Appl Microbiol. 1999;86:331–336. doi: 10.1046/j.1365-2672.1999.00674.x. [DOI] [PubMed] [Google Scholar]
- Silva N, Junqueira VCA, Silveira NFA. Manual de Métodos e Análise Microbiológica de Alimentos. 2nd ed. Livraria Varela; São Paulo: 1997. p. 317. [Google Scholar]
- Vender RJ, Marignani P. Salmonella colitis presenting as a segmental colitis resembling Crohn’s disease. Dig Dis Sci. 1983;28:848–851. doi: 10.1007/BF01296909. [DOI] [PubMed] [Google Scholar]