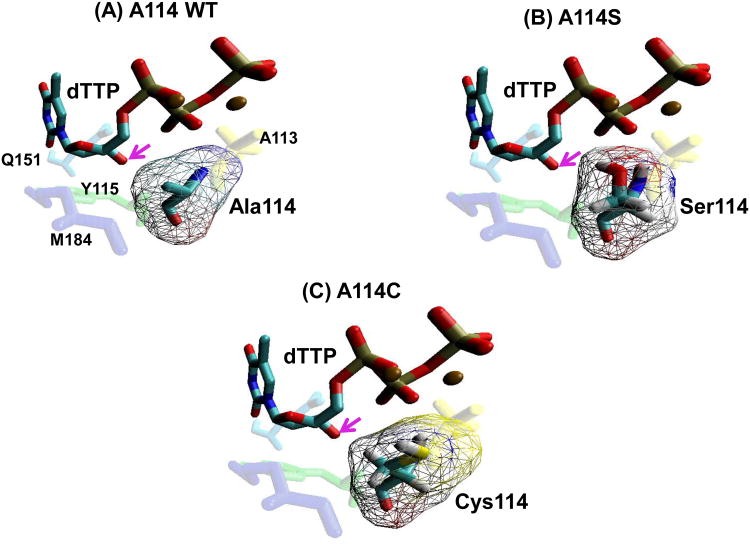
Figure 1. Structural modeling of the HIV-1 RT residue 114 mutants at the dNTP-binding pocket.
(A) The position of the HIV-1 RT A114 residue (teal) in relation to an incoming dTTP substrate (blue/red), and Y115 (green), A113 (yellow), M184 (purple) and Q151 (light blue), is shown using coordinate set 1rtd.pdb from the Protein Data Bank (Huang et al., 1998). (B) and (C) The positions of the S114 and C114 mutant residues were constructed by the program Pymol (Schrödinger) that manually selected the optimal rotamer for serine or cysteine at residue 114 of HIV-1 reverse transcriptase from Huang et-al 1999 (Huang et al., 1998). These were then minimized with AMBER 9.0 with 500 cycles (250 steepest descent) with implicit solvent. Arrows indicate the 3′ OH of the incoming dTTP. Volume of amino acids were calculated as described previously (Zamyatnin, 1984).