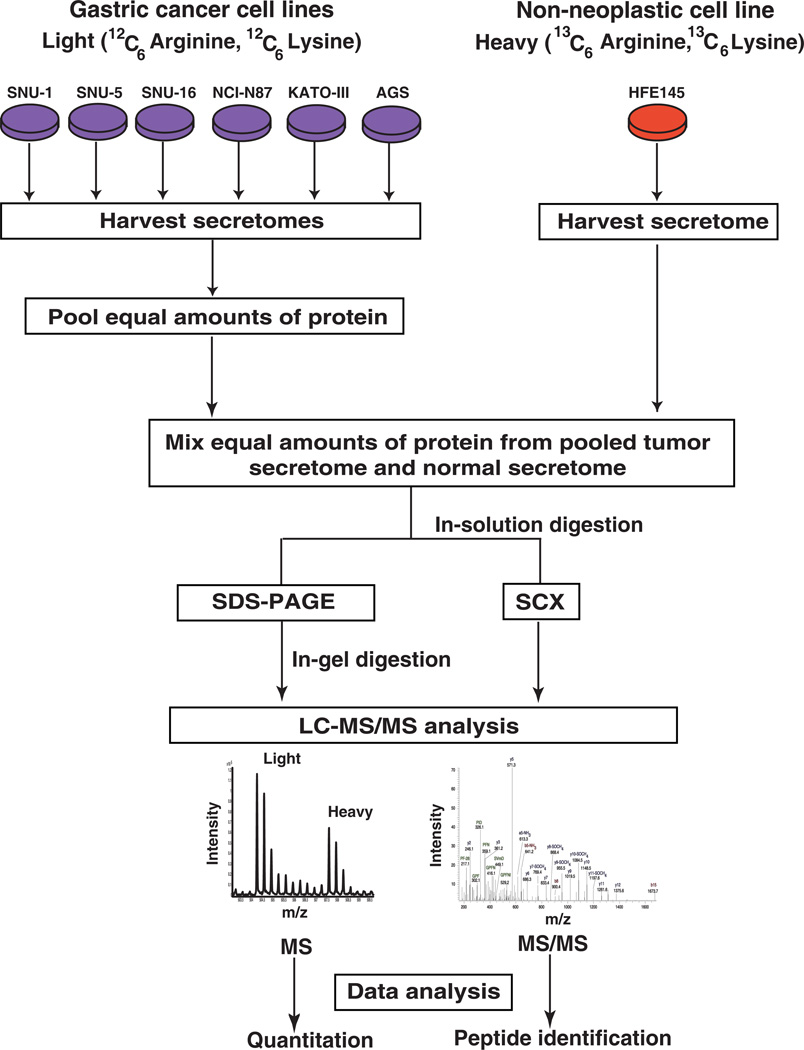
Figure 1. Workflow employed for the analysis of gastric cancer secretome.
SILAC was employed to identify the more abundant proteins in the secretome of gastric cancer cells compared to the normal cells. Gastric cancer cell lines (AGS, SNU-1, SNU-1, SNU-5, SNU-16, NCI-N87, AGS and KATO-III) were grown in medium containing normal arginine and lysine. The normal gastric epithelial cell line was grown in a medium containing heavy arginine and lysine. Equal amounts of protein from pooled tumor secretome and from normal secretome were mixed. Proteins were subjected to fractionation by SDS-PAGE and strong cation exchange chromatography. The fractionated proteins were subjected to LC-MS/MS analysis using an LTQ-Orbitrap Velos mass spectrometer. The data was searched and quantitated using the Proteome Discoverer software suite.