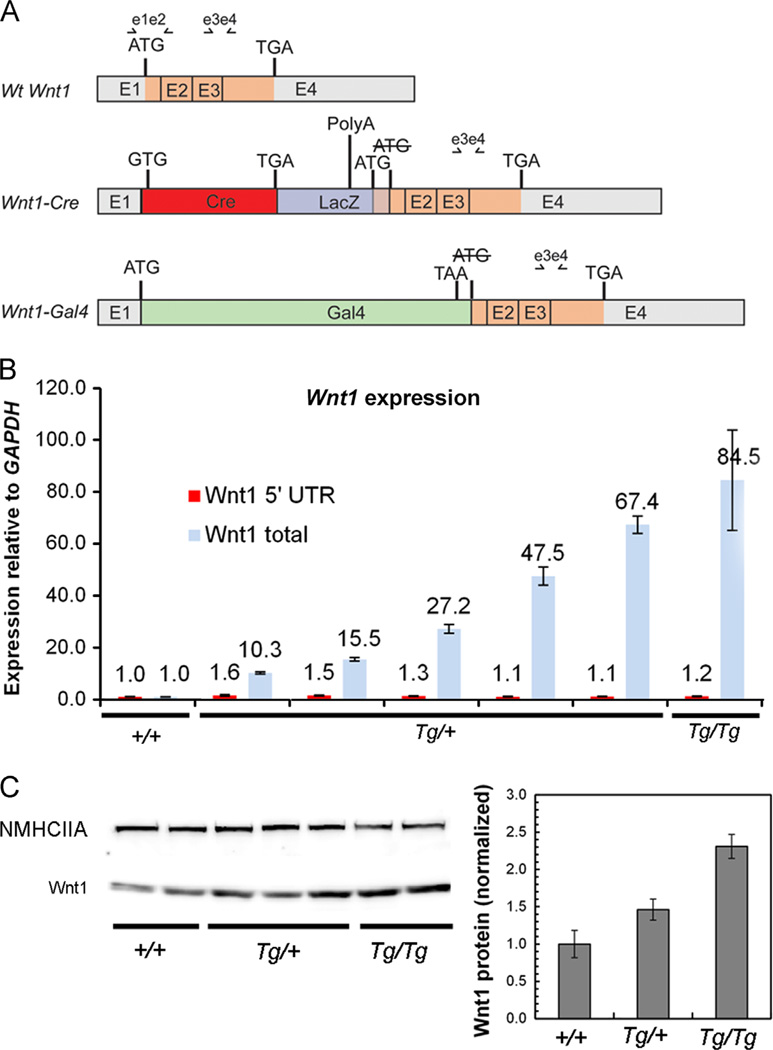
Fig. 2.
Molecular basis for ectopic Wnt1 expression in Wnt1-CreTg embryos. (A) Schematic representation of wild-type Wnt1 transcript and two Wnt1-CreTg-specific Wnt1 transcripts. A Wnt1-Cre chimeric transcript carrying the complete Cre open reading frame, followed by part of the LacZ gene, also has an open reading frame predicted to encode the entire Wnt1 transcript plus 165 5′ base-pairs derived from the LacZ sequence. The Wnt1-Gal4 chimeric transcript is not predicted to encode a complete Wnt1 open reading frame. (B) Quantitative RT-PCR using primers recognizing all three Wnt1 transcripts (e3e4 in A) indicates variable overexpression of Wnt1 transcript in E13.5 embryonic Wnt1-CreTg heads. Quantitative RT-PCR using primers recognizing only the wild-type Wnt1 transcript indicate that endogenous Wnt1 expression remains similar between genotypes (e1e2 in A). Error bars represent standard deviation from qRT-PCR performed on individual embryos in triplicate. (C) Western blot analysis of Wnt1 from E14.5 embryonic head extracts reveals a 1.5-fold upregulation of Wnt1 protein in Wnt1-CreTg/+ embryos, and an approximately 2.3-fold upregulation of Wnt1 protein in Wnt1-CreTg/Tg embryos. Non-muscle myosin heavy chain IIA was used as a loading control, and error bars represent standard error.