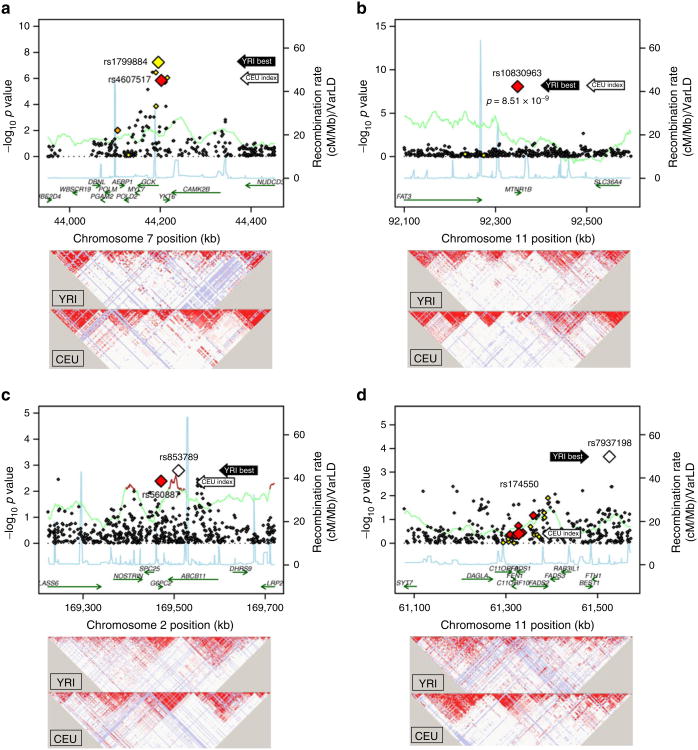
Fig. 1.
500 kb regional association plots centred at the index SNPs identified from EuA samples at GCK, MTNR1B, FADS1 and G6PC2. The x-axis denotes genomic position and the y-axis denotes the –log(p value) for each SNP. The two larger-sized data points represent the best SNPs within the region in the AfA CARe sample (black arrows) and the index SNPs (white arrows, locating the centre of the region) previously reported from EuA population samples. The colour of each data point indicates its LD value (r2) with the index SNP based on HapMap 2 YRI: white, r2=0.0–0.2; yellow, r2=0.2–0.5; orange, r2=0.5–0.8; red, r2=0.8–1.0. The blue line represents the recombination rate. The green line shows the varLD score at each SNP and is highlighted with dark brown if the varLD score is ≥95th percentile of the genome-wide varLD score, comparing LD information between YRI and CEU HapMap 2 samples [29]. (a)GCK region: the index SNP rs4607517 has a p value of 1.42 × 10-6 in AfA CARe individuals, while the best SNP rs1799884 has a p value of 5.79 × 10-8. Their r2 values are 0.469 in HapMap 2 YRI and 1.0 in HapMap 2 CEU. (b) MTNR1B region: rs10830963 is the index SNP and the best SNP within the region in AfA CARe individuals, with a p value of 8.51 × 10-9. (c) G6PC2 region: the index SNP rs560887 has a p value of 4.08 × 10-3 in AfA CARe individuals and the best SNP rs853789 has a p value of 1.62 × 10-3. The r2 value is not known in HapMap 2 YRI and is 0.692 in HapMap 2 CEU. A known functional SNP in the region, rs13431652 [38], has r2=0.923 with rs560887 and r2=0.595 with rs853789 in HapMap 2 CEU, but in HapMap 2 YRI the r2 of rs13431652 with rs853789 is 0.004. (d) FADS1 region: the index SNP rs174550 has a p value of 0.399 in AfA CARe individuals and the best SNP rs7937198 has a p value of 2.22 × 10-4. Their r2 values are 0.00 in HapMap 2 YRI and 0.24 in HapMap 2 CEU