Abstract
Lanthanide-induced enhancement of the longitudinal relaxation of nitroxide radicals in combination with orthogonal site-directed spin labeling is presented as a systematic distance measurement method intended for studies of biomacromolecules and biomacromolecular complexes. The approach is tested on a water soluble protein (T4-lysozyme) for two different commercially available lanthanide labels, and complemented by previously reported data on a membrane inserted polypeptide. Single temperature measurements are shown to be sufficient for reliable distance determination, with an upper measurable distance limit of about 5-6 nm. The extracted averaged distances represent the closest approach in LnIII-nitroxide distance distributions. Studies of conformational changes and of biomacromolecule association-dissociation are proposed as possible application area of the RE-based distance measurements.
Keywords: EPR, proteins, lanthanides, nitroxide radicals, nanometer-range distances
1 Introduction
Addressing biochemical questions usually requires a combination of multiple experimental tools, which verify and supplement each other. While X-ray crystallography and NMR spectroscopy are generally recognized as the major methods to obtain atomic-resolution structures of proteins (or other biomacromolecules), a number of spectroscopic tools has been developed to access further information on protein dynamics and folding, conformational changes, agglomeration, protein-cofactor and protein-protein interactions. To give a few examples in optical spectroscopy, surface plasmon resonance[1, 2] or single molecule fluorescence,[3, 4] in particular Förster resonance energy transfer,[5, 6] can be mentioned. A particular strength of optical techniques is in observing dynamics on a single molecule level. On the other hand, electron paramagnetic resonance (EPR) spectroscopy, a sister technique of NMR, provides the most accurate long range distance measurement approaches. The EPR distance measurements are typically based on the pulse double electron-electron resonance (DEER or PELDOR) experiment[7, 8] in its dead-time free version.[9, 10] The EPR approaches do not require sample crystallization, they are not limited by the biomolecule size, and can be applied to soluble and membrane inserted biomolecules.[11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21] Long range EPR-based distance measurements can efficiently supplement X-ray diffraction[22] or NMR.[23] EPR can also be used as a stand-alone technique, especially in studies of conformational changes of large biomolecules.[24, 25]
As most biomacromolecules are diamagnetic, the application of EPR techniques requires spin labeling. On proteins this is usually done by attaching nitroxide spin labels with thiolreactive groups to engineered cysteine residues.[26, 27, 28] The advantage of such an approach is site selectivity and, as a consequence, safe assignment of the measured distance with respect to the primary structure of the biomacromolecule. The disadvantage is the relatively low information content: only one intramolecular distance can usually be accessed per sample.
To enhance the accessible information per sample, one can use spectroscopically orthogonal spin labels.[29, 30, 31, 32, 33, 34, 35, 36] In particular, the GdIII-nitroxide DEER experiment has a very good performance in distance measurements and, importantly, it is possible to selectively detect each of the two types of spin labels.[32] Based on this spectroscopic selectivity, it was demonstrated that combining GdIII-chelate spin labels and nitroxide spin labels allows for selective detection of GdIII-nitroxide, GdIII-GdIII and nitroxide-nitroxide distances in the same sample,[34, 35, 36] which can be used to simultaneously access different intramolecular distances[34] or to obtain information on biomolecule oligomerization or aggregation.[36] The local environment of individual spin labels can be monitored at the same time.[36] These advantages can become crucial in studies of biomolecule aggregation, protein-cofactor and protein-protein interactions or association-dissociation of biomolecular complexes.
In this paper we discuss another lanthanide-based approach for distance measurements in EPR, based on the enhancement of nitroxide radical relaxation in the presence of fast relaxing lanthanide ions.[37, 38, 39] While several EPR studies were reported, where fast relaxing agents were naturally present in biomacromolecules,[40, 41, 42, 43, 44] the site-directed introduction of EPR relaxation agents and corresponding distance measurements have not yet been examined in detail. This approach is similar to the paramagnetic relaxation enhancement (PRE) approach in NMR, but due to the larger magnetic dipole moment of nitroxide radicals, as compared to any magnetic nuclei, the relaxation enhancement (RE) approach in EPR might access longer interspin distances than PRE. Importantly, stochastic and static dipolar interactions manifest themselves differently and thus require quite different pulse experiments for their detection. While in DEER distance measurements the upper distance limit is determined by the transverse relaxation time T2 of the observer species, it is limited by the longitudinal relaxation time T1 in RE techniques. As a result, RE experiments can potentially be combined with the DEER-based distance measurements, thus further increasing the available information content. Development of lanthanide-based techniques in NMR, such as PRE or pseudo-contact shift measurements, where important contributions were made by Ivano Bertini and coworkers,[45, 46, 47, 48, 49] allows one to suggest a combination of EPR-based and paramagnetic NMR-based long range distance measurements as a possible future tool in structural biology.
Methods for lanthanide labeling benefited from the development of paramagnetic NMR techniques.[50, 51] On the other hand, orthogonal labeling with both lanthanide ions and nitroxide radicals is much less developed. If non-identical biomolecules can be labeled prior to their association into a complex, orthogonal labeling can be a relatively easy task. For smaller monomeric molecules one or more paramagnetic centers can be synthetically built.[36, 39] Specific properties of a certain protein, as, for instance, protection of a labeling site by substrate binding, could be used for orthogonal labeling.[52] An approach that uses unnatural amino acids for nitroxide labeling[53] and a thiol-specific reaction for lanthanide labeling is currently the only tested systematic protocol of orthogonal chemo-selective spin labeling of large monomeric protein molecules.[54] While the same orthogonal labeling approach is used in this work, it is worth mentioning that further advances in the area of chemo-selective orthogonal spin-labeling can make spectroscopically orthogonal distance measurements in EPR even more versatile.
This work is based on the detailed spectroscopic investigation of the RE-based distance measurement technique on a different model system.[39] Here we aim to establish the RE approach as a systematic distance measurement tool in bio-EPR applications by addressing problems that arise in potential routine application of the approach. In particular, we discuss how to reduce the effort required to measure a single distance. Performance of the RE approach for membrane inserted[39] and soluble (this work) biomolecules is compared. The optimal experimental conditions, type of obtained distance constraints, and potential applications of the RE approach are discussed.
The ability of lanthanide ions to substitute each other in chelate complexes[55] allows for calibrating lanthanide-induced RE distance measurements against GdIII-nitroxide DEER. This approach has been already used in our study of membrane inserted synthetic WALP23 polypeptides.[36, 39] In this work we analyze RE in orthogonally labeled water-soluble T4-lysozyme, for which GdIII-nitroxide DEER data were recently reported.[54]
1.1 Extraction of Distance Information from Longitudinal RE Data
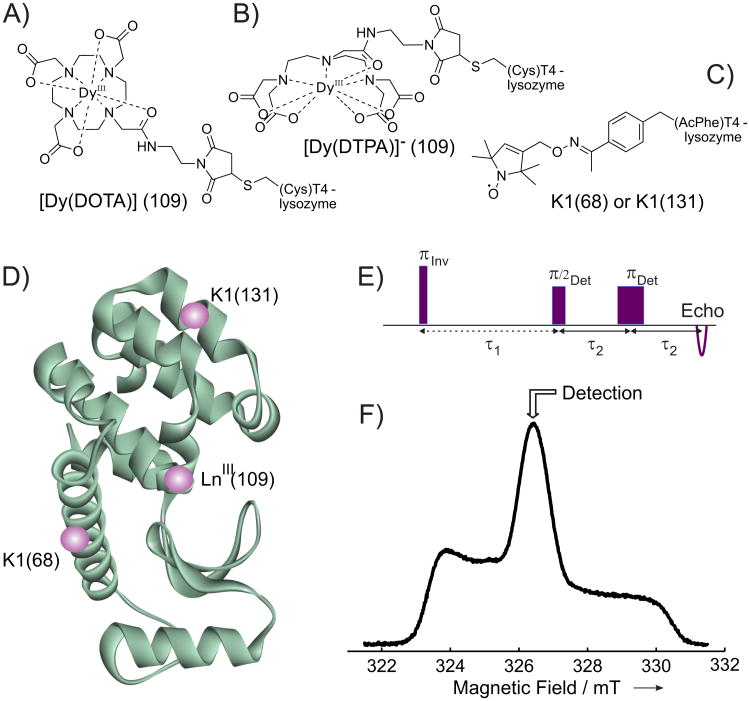
The shape of the nitroxide echo-detected (ED) EPR spectrum and the detection positions in the inversion recovery (IR) experiment are shown in Figure 1.
Figure 1.
Chemical structures of the side chains formed after attachment of A) [Dy(DOTA)], B) [Dy(DTPA)], and C) nitroxide radical (K1 side chain) to the T4-lysozyme. D) Ribbon model of wild-type T4-lysozyme (PDB ID code 1L63) with LnIII label at position 109 and nitroxide side chain K1 either at position 68 or 131 (spheres are placed approximately at positions of paramagnetic labels). E) IR pulse sequence used in the RE experiments. F) Nitroxide ED EPR spectrum for the sample 68K1-109[Dy(DTPA)] measured at 80K. The detection position for the RE experiment is marked with an arrow.
The RE value Δk is defined as the difference of the inverse relaxation times of a slowly relaxing species in the presence T1s and absence of a fast relaxing species T1s,0.
| (1) |
In the data processing we use factorization [38, 56] of a longitudinal magnetization decay trace into a product of a non-perturbed decay V0(t) and an additional DyIII-induced RE-decay Vk(t):
| (2) |
This factorization is exact for a single type of slowly relaxing species and it is a fairly good approximation in case of the distribution of non-perturbed longitudinal relaxation times.[39]
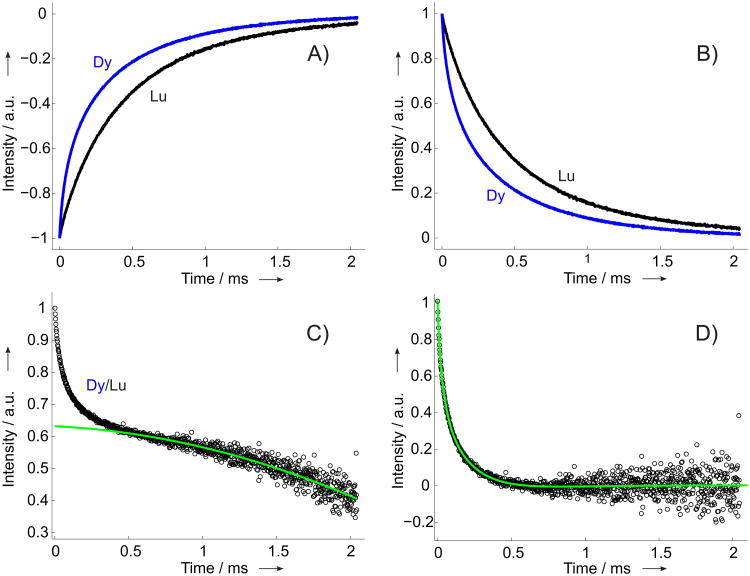
The main data processing steps are illustrated in Figure 2. The primary (IR) times traces were measured until 1% of their initial value and rescaled to the standard interval [-1,0] by reference measurements of the echo intensity in the absence of the inversion pulse, c.f. Figure2(A,B). For each particular nitroxide label site the IR time trace of the DyIII-labeled sample was divided by the corresponding time trace of a LuIII-labeled sample, detected at the same temperature, c.f. Figure 2(C). The resulting RE time trace contained contributions from intra- and intermolecular RE. The intramolecular contribution is responsible for the fast initial decay of the signal. In an ideal situation of 100% DyIII labeling this contribution would lead to the signal decay down to zero, whereas in the presented experimental series we observed decays down to values ≈(0.6 − 0.8), indicating incomplete DyIII labeling. The remaining signal comes from molecules not labeled with a DyIII and thus reflects the intermolecular RE. This slowly decaying intermolecular contributions were fitted by low order (1st to 3rd) polynomial functions and subtracted from the RE traces. The final intermolecular RE time traces were rescaled and fitted by multiexponential fit curves to extract 1/e times that were used as a measure of the average relaxation enhancement . For a single DyIII-nitroxide distance it has been shown[38] that summing up all the orientations of the DyIII center and neglecting the weak g- and A-anisotropy of nitroxide radicals results in an average 1/e time τ1 and corresponding RE value that scales with the inverse 6th power of the interspin distance:
Figure 2.
Illustration of the data processing procedure for sample pair 131K1-109[Dy(DOTA)]/131K1-109[Lu(DOTA)]. IR traces were measured at X band at T = 80K. A) Primary IR data, renormalized to the interval [-1,0] by reference measurements with deactivated inversion pulse (blue - DyIII-loaded, black - LuIII-loaded). B) The two IR traces from A) inverted. C) The RE time trace, obtained by division of the renormalized IR traces (black) and the background fit function (green). D) The background corrected intramolecular RE trace (black) and the multiexponential fit function (green), used for extracting the 1/e time.
| (3) |
The anisotropies of resonance frequency for the fast (Dy) and slowly (nitroxide) relaxing species enter the averaging in Equation (3). While the anisotropy for DyIII centers is strong enough and is thus taken into account in the averaging procedure, the anisotropy for nitroxide radicals is rather weak and can be safely neglected. The result of RE measurements is thus essentially independent on the detection position within the nitroxide spectrum. Note also that no correlation between the orientations of nitroxide and DyIII labels was observed in the reference GdIII-nitroxide DEER experiments.[54]
For a sample with a distribution of DyIII-nitroxide distances the 1/e time obtained according to Equation (3) corresponds to an averaged distance which somewhat underestimates the actual mean distance extracted from the reference GdIII-nitroxide DEER measurements.[39] Our T4-lysozyme data presented in the Results section confirm this observation. The averaged relaxivity C̄ (T1f) can be fitted to the temperature dependent RE data, assuming a single T1f time for all DyIII species with a power-law dependence on the temperature:[38, 39]
| (4) |
While Equation (4) is phenomenological and does not give direct insight into the dominating Dy relaxation mechanism, it suffices for the description of RE data in the relevant (limited) temperature range.[38, 39] The parameters Tmax and p determine the maximum RE temperature and the width of the bell-like RE temperature dependence, but do not affect the vertical scaling of the RE curve. The DyIII-nitroxide distance, in turn, exclusively affects the vertical scaling and has little effect on the shape of the RE curve.[39] Still, some change of the RE curve was observed for samples with a large fraction of DyIII-nitroxide distances below 2 nm, for which the distribution of T1s times should be particularly broad. [39] Note also the 10-15% precision of the values and the smooth maximum of the RE temperature dependence. This results in rather large uncertainty for the Tmax and p. The parameter A can be computed from the detection frequency and from the g-tensor of DyIII centers. A typical value for A at X band for DyIII -nitroxide pairs is ∼ 10−11 s. Variation of A leads to some mismatch between the actual maximum RE temperature and the parameter Tmax, but does not affect the following distance determination. The fitted values of Tmax, p and A for all four samples are listed in Table 1.
Table 1.
Simulation parameters for RE curves, according to Equation (4).
| Sample | A [10−11 s] | Tmax [K] | p |
|---|---|---|---|
| 68K1-109[Dy(DOTA)] | 1.0 | 90 | 2.4 |
| 68K1-109[Dy(DTPA)] | 1.0 | 85 | 2.4 |
| 131K1-109[Dy(DOTA)] | 1.0 | 80 | 3.5 |
| 131K1-109[Dy(DTPA)] | 1.0 | 80 | 3.4 |
The g-tensor of DyIII species determines their resonance frequency at each orientation with respect to the external magnetic field. The magnitude of RE depends on the resonance frequencies of fast and slowly relaxing spins. In the RE value ( ) the angular dependence is averaged out, but is still dependent on the eigenvalues of the DyIIIg-tensor. The CW EPR spectra of [Dy(EDTA)] and [Dy(DOTA)] complexes were measured experimentally and in both cases the spectra are consistent with an axial g-tensor with g‖ ≈ 15 and g┴ ≈ 4.2.[39, 57] Due to the chemical similarity between EDTA, DTPA and DOTA and because for two of these chelators corresponding g-tensors seem to be very similar, we assumed in calculations an axial g-tensor with g‖ ≈ 15 and g┴ ≈ 4.2 for [Dy(DOTA)] and for [Dy(DTPA)]. It was also tested that rather large variations of the g-values produce very small deviations in computed RE.[38]
2 Results
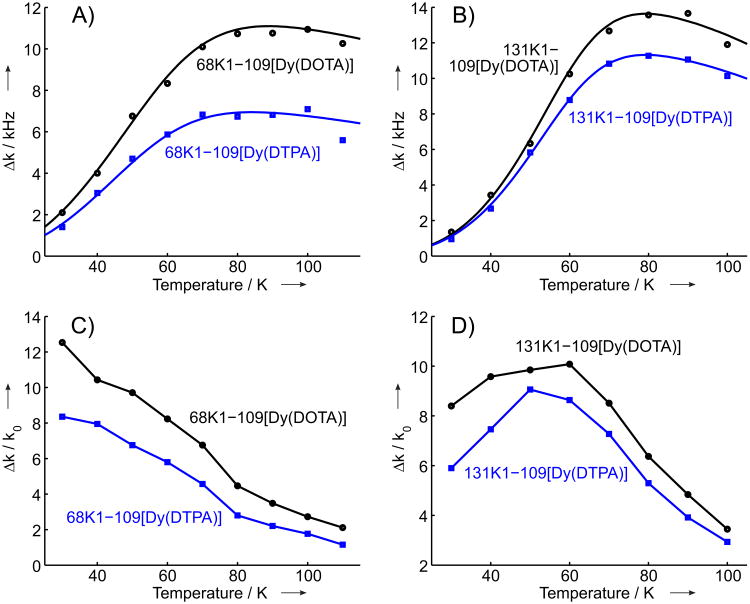
Temperature dependencies of DyIII-induced RE for the two studied mutants and for both types of DyIII labels are shown in Figure 3. The corresponding extracted distances are compared to the reference GdIII-nitroxide DEER data[54] in Table 2. For GdIII-nitroxide DEER the mean distances as well as the widths of the distance distributions are given, whereas the RE approach currently allows only for the determination of an average distance. The relative contributions of the intramolecular RE are also given in Table 2 (in percents). One can see that the DyIII-labeling efficiency varied between 18% and 34%, in a fairly good agreement with previously estimated GdIII labeling efficiencies on the same T4-lysozyme mutants.[54]
Figure 3.
Temperature dependent RE (Δk) and RE contrast (Δk/k0, dimen-sionless) for the four studied T4-lysozyme samples: A) RE for the samples 68K1-109[Dy(DOTA)] (black circles) and 68K1-109[Dy(DTPA)] (blue squares), corresponding solid lines show the simulations of RE temperature dependencies (see text). B) RE for the samples 131K1-109[Dy(DOTA)] (black circles) and 131K1-109[Dy(DTPA)] (blue squares), corresponding solid lines show the simulations of RE temperature dependencies (see text). C) RE contrast for the samples 68K1-109[Dy(DOTA)] (black) and 68K1-109[Dy(DTPA)] (blue). D) RE contrast for the samples 131K1-109[Dy(DOTA)] (black) and 131K1-109[Dy(DTPA)] (blue).
Table 2.
Comparison between distances obtained from X-band relaxation enhancement and GdIII - NO measurements.
| Sample | RE | DEER (r ± ξ)1 | DyIII labeling2 |
|---|---|---|---|
| 68K1-109[Dy(DOTA)] | 2.96nm | (3.5 ± 0.2 )nm | 26% |
| 68K1-109[Dy(DTPA)] | 3.20nm | (3.5 ± 0.3 )nm | 18% |
| 131K1-109[Dy(DOTA)] | 2.86nm | (3.5 ± 0.8 )nm | 34% |
| 131K1-109[Dy(DTPA)] | 2.95nm | (3.5 ± 0.7 )nm | 33% |
Mean distance (r) and FWHM (2ξ = FWHM) from GdIII-nitroxide DEER.[54]
Averaged over all RE traces at different temperatures.
The RE values obtained with [Dy(DTPA)] labels are systematically lower than the corresponding RE values for [Dy(DOTA)] labels. The difference is somewhat larger for the mutant with the K1 side chain at position 68, where the corresponding difference in obtained distances is about 0.2 nm. For K1 at site 131 the averaged DyIII-nitroxide distance for the [DyIIIDOTA]-labeled sample is only about 0.1 nm shorter than for the [DyIIIDTPA]-labeled sample. [Dy(DOTA)] and [Dy(DTPA)] labels have three and four carboxylic groups respectively (one carboxylic group is used in each case to attach the chelator to the maleimido linker, Scheme 1). Thus, the [Dy(DOTA)] label is neutral whereas the [Dy(DTPA)] label is negatively charged. This might slightly change the position of the DyIII chelate complex with respect to the protein and create the difference in the obtained DyIII-nitroxide distances. Slight differences between [Gd(DOTA)]- and [Gd(DTPA)]- labeled samples were observed as well in the corresponding GdIII-nitroxide DEER experiments.[54]
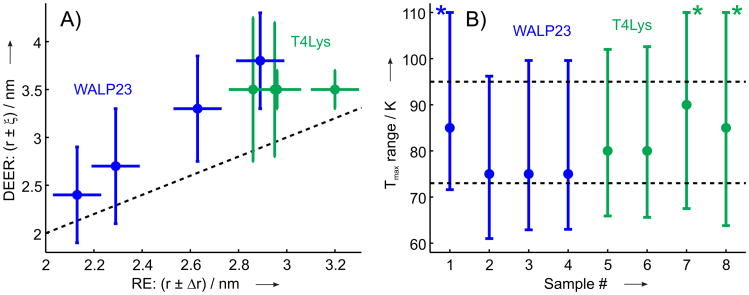
The RE contrast is defined as the ratio of the averaged RE and the inverted longitudinal relaxation time (k0 = 1/T1s,0) of nitroxide raticals in the absence of the fast relaxing agent. This value, measured for a particular DyIII-nitroxide distance, allows estimation of the longest accessible distances for the given label pair, since a minimum RE contrast of ∼ 0.5 is required for reliable RE measurement,[39] and since for a given k0 the RE contrast has the same distance dependence as the value. Taking into account the RE contrasts at 80 K for the two studied mutants (Figure 4) and the obtained distances of about 3 nm, as well as the limiting contrast of /k0 ≈ 0.5 one can conclude that distances up to ∼ 5 nm should be accessible with this method. This estimate is close to the one obtained with the membrane spanning α-helical WALP23 polypeptide.[39]
Figure 4.
A) Comparison between distances obtained from DyIII-induced RE and GdIII-nitroxide DEER measurements.[36, 54] For RE measurements the distances are given with estimated error bars, for GdIII-nitroxide DEER measurements mean distances with ±ξ bars are plotted, compare to Table 2. B) Temperature region for 90% of maximum value Δkmax for the four T4-lysozyme samples and four WALP23 samples studied previously,[39] circles show temperatures of maximum RE. In the cases, when the upper temperature limit of the region for 90% of maximum value Δkmax was above 110K, corresponding data are marked with an asterisk. In both plots data are shown for T4-lysozyme in green and for WALP23 in blue.
3 Discussion
While labeling efficiency of LnIII labels is not so easy to assess from GdIII-nitroxide DEER measurements,[54] it can be directly extracted from the LnIII-induced RE data (Figure 2 and Table 2). By combining RE experiments with GdIII-nitroxide DEER to measure the nitroxide labeling efficiency, one can selectively determine the labeling efficiencies for both types of spin labels. This type of measurements offers a useful flexibility in association-dissociation experiments, especially in the case of excess of some type of complex-forming biomolecules.
The use of LuIII-labeled samples for diamagnetic reference in this study was mainly driven by the easy dimerization of T4-lysozyme. In a general case additional diamagnetic labeling is not required, because typically the presence or absence of a diamagnetic label at some remote site does not induce any measurable change on the relaxation of nitroxide spin labels. Thus, the singly nitroxide labeled sample before the DyIII labeling could be used as a diamagnetic reference, and preparation of a separate reference sample could be avoided.
The distance range accessible by the presented method is similar to that of DEER on membrane exposed nitroxide radicals,[20] but it is reduced compared to the distance range accessible by DEER for soluble proteins, especially in deuterated solvents.[58] Importantly, the RE-accessible distance range appears to be rather stable with respect to the nitroxide environment: the values obtained for the DyIII-nitroxide distance of about 3 nm for water soluble T4-lysozyme (between 6 kHz and 12 kHz, Figure 3) and for membrane-incorporated WALP23 polypeptide (∼10 kHz)[39] are rather similar. While for the nitroxide K1-side chain at site 68 the RE contrast at the maximum of RE curve (T ∼ 80 K) is about 4, the sample with the K1 side chain at site 131 shows a RE contrast of about 6 at 80 K. The optimum RE contrast was observed at about 50-60 K for K1 at site 131 and at the lowest measured temperature (30 K) for K1 at site 68 (Figure 3). For K1 at site 68 the RE temperature dependence had a broader maximum compared to the sample with K1 at site 131; this might explain the change in the RE contrast curve. Note that a significant part of this discrepancy could be accounted for by the uncertainty of the measurements. For the membrane inserted WALP23 the RE temperature dependencies were similar to the one observed for K1 at site 131 (Figure 3) and the optimum RE contrast temperature did not depend on the nitroxide site.[39]
Further increase of the accessible distance range could be possible if for any other LnIII ion the optimum RE is achieved at lower temperatures, where the non-perturbed longitudinal relaxation of nitroxide radicals is slower and thus a smaller RE value is required for the limiting RE contrast of 0.5. On the other hand, for DyIII labels the optimum RE temperature is in the range between 80 and 90K, thus the distance measurements could be performed with liquid nitrogen cryostats and do not necessarily require use of liquid helium. Longitudinal relaxation of nitroxide radicals can also be tuned, for instance, by substituting the geminal dimethyl groups by spirocyclohexyl moieties.[59] Such modifications of nitroxide radicals could lead to improved RE contrast and, thus, to a broader accessible distance range.
The DEER experiment is capable of measuring distance distributions, whereas the RE method in its current state of development provides only averaged distances. Yet, while we only made single distance extraction in this study, the measurement of two sufficiently different distance peaks or estimating the width of a broad distance distribution might be feasible with the RE experiment.
Based on the RE data obtained on membrane inserted WALP23 polypeptides[39] ([Dy(DOTA)] labels) and on orthogonally labeled water soluble T4-lysozyme ([Dy(DOTA)] and [Dy(DTPA)] labels, this work), one can draw some preliminary conclusions on the performance of the RE-based distance measurements. Figure 4(A) correlates distances obtained from GdIII-nitroxide DEER and from DyIII-induced RE. The presented correlation reveals that RE measurements underestimate the actual LnIII-nitroxide distances and that the deviation between RE-based distance and the actual mean distance tends to increase for longer LnIII-nitroxide distances. Such a behavior implies some underlying physical mechanism, which could be speculatively attributed to an indirect pathway relaxation enhancement, most probably via the acceleration of proton relaxation in the vicinity of paramagnetic centers.[39, 60] Distance information with such reduced precision might still be of interest for monitoring conformational changes of biomacromolecules or association/dissociation events. A calibration of RE distances against reference DEER data might further improve the precision of these distance measurements and allow for the direct use of RE data as quantitative distance constraints.
As the distance is extracted exclusively from the vertical scaling of the RE curve, it is not necessary in routine applications to measure the entire temperature dependence. Figure 4(B) shows temperature ranges, where RE exceeds 90% of its maximum value. This corresponds to 1-2% underestimation of the DyIII-nitroxide distance, which can usually be tolerated, since other uncertainties in interpretation of distances are larger. One can see that a rather broad temperature range is available in this case and that temperatures between 75K and 85K are always close to the maximum of RE temperature dependence. This implies that for a distance measurement with practically sufficient precision, RE data at a single temperature are sufficient. From the presented set of data, the temperature of 80K can be recommended for such single point RE measurements.
4 Conclusions
The presented RE-based distance measurement approach is systematically applicable and does not rely on the presence of native fast relaxing centers in the biomolecule under study. The experimental evidence obtained so far indicates that the RE-based approach is rather robust, can be performed as a single temperature measurement, and works well in a broad labeling efficiency range. The results indicate that the approach will be applicable in studies of protein association-dissociation or conformational changes of proteins/protein complexes. With the same labeling both RE and DEER distance measurements can be performed. Since sample production requires much greater effort than the spectroscopic measurements, both RE and DEER experiments should be performed in application work to benefit from complementary information.
In the present work we only extracted a single closest approach distance from RE time traces. Due to a strong change of RE with the LnIII-nitroxide distance, extraction of more detailed distance information might be feasible, most probably with lower resolution than in DEER experiments. Work along these lines is currently in progress.
5 Experimental Section
5.1 EPR Measurements
Pulsed EPR measurements were carried out at X band (9.3 - 9.4 GHz) on a Bruker ElexSys 580 spectrometer equipped with a Bruker split-ring resonator (ER 4118× - MS3). Temperature stabilization was performed with a continuous flow He cryostat (ESR900, Oxford Instruments) equipped with an Oxford Instruments temperature controller ITC 503S. A temperature range from 30K to 100K was covered in 10K steps. The relaxation enhancement (RE) data were obtained from T1 relaxation measurements with an inversion recovery (IR) pulse sequence (Figure 1(E)) as described earlier [38, 39] with τ1 = 500ns, an inversion pulse of 12 ns and the Hahn echo detection sequence with 52(π/2)-104(π)ns pulses and interpulse delay τ2 = 200 ns. The inversion pulse was set to the largest reproducibly possible bandwidth to minimize the effect of spectral diffusion on the measured IR trace. It was shown that already with an inversion pulse of 32 ns a fairly good agreement between IR data and, presumably spectral-diffusion-free, saturation recovery data could be achieved.[38] Furthermore, only minor effects on T1 were observed in recent work comparing full inversion of the nitroxide spectrum by an adiabatic passage pulse to partial excitation by a rectangular pulse.[61] Echo-detected (ED) EPR spectra were recorded with a 16(π/2)-32(π)ns Hahn echo sequence with an interpulse delay of 200 ns.
5.2 Sample Preparation
The sample preparation procedure combines previously established protocols.[53, 54, 60]
Chemicals
The following acronyms will be used: DOTA, 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid; DTPA, diethylene triamine pentaacetic acid; DTT, dithiothreitol; EDTA, ethylenediaminetetraacetic acid; MOPS, 3-(N-morpholino)propanesulfonic acid; Xylenol Orange, 3,3′-Bis[N,N-bis(carboxymethyl)aminomethyl]-o-cresolsulfonphthalein disodium salt.
Unless stated otherwise, all commercially available chemicals were used as received. Maleimido mono-amide DOTA·CF3COOH·HPF6 and maleimido mono-amide DTPA·3CF3COOH were purchased from Macrocyclics, Dallas. Anhydrous DyCl3 (99.99%), DTT (99%), MOPS (99.5%), sodium acetate (>99%) were purchased from Sigma-Aldrich. EDTA (>99%) was purchased from Fluka. Glycerol (99%) was purchased from Honeywell. NaCl (99.5%) was purchased from J.T.Baker. Xylenol Orange was purchased from TCI.
A MOPS (50mm ), NaCl (500 mm ), glycerol (10% v/v), pH 6.8 buffer was used for all samples. This buffer was shown to strongly reduce the non-specific binding of LnIII complexes to the surface of T4-lysozyme.[54]
LnIII-complex formation
Maleimido mono-amide (39.3mg, 0.05mmol, 1 equiv.) DOTA·CF3COOH·HPF6 was dissolved in buffer (2.5 mL) and DyCl3 or LaCl3 aqueous solution (347 μL, 0.05 mmol, 1 equiv.) was added, forming [Ln(DOTA)] complex. The solution was stirred for 24 h in the dark. Un-chelated LnIII ions were determined by eye detection with xylenol orange using a modified published procedure[62] and subsequently sequestrated with the necessary amount of EDTA. The LnIII-complex solution (100 μL) was diluted with acetate buffer solution (1.2 mL, 50 mm, pH = 5.8) and few drops of xylenol orange solution (0.016 mm) were added. Upon adding a few drops of EDTA in water (10 mm), the solution color turned from red into orange indicative of the disappearance of LnIII aquo-ions. For [Ln(DTPA)] complex formation maleimido mono-amide DTPA·3CF3COOH ((28.2 mg, 0.03 mmol, 1 equiv.)) was dissolved in buffer (1 mL) and LnCl3 aqueous solution (228 μL, 0.03 mmol, 1 equiv.) was added. The solution was treated as previously described for DOTA.
Protein Preparation and Labeling
Protein preparation and nitrox-ide labeling were performed according to Fleissner et al.,[53] and lanthanide labeling was performed according to Garbuio et al.[54] The DyIII-nitroxide orthogonal labeling approach was tested on two double mutants of bacteriophage T4-lysozyme with a genetically encoded unnatural amino acid p-acetyl-L-phenylalanine (p-AcPhe) either at site 68 or at site 131 and a cysteine at position 109. The acetyl group of the residue p-AcPhe was reacted with the hydroxylamine pendant of a nitroxide radical to give rise to a ketoxime-linked nitroxide side chain K1 (Figure 1). In each mutant the cysteine at site 109 was used for binding of the DyIII-based spin label. Two different commercially available functionalized chelators, maleimido-monoamide DOTA and maleimido-monoamide DTPA, were used to efficiently trap DyIII ions. The resulting chelate complexes, [Dy(DOTA)] and [Dy(DTPA)]− were then covalently attached to the protein through the chemo-specific reaction between the maleimido moiety and the cysteine sulfhydryl (Figure 1).
A T4-lysozyme sample (30 μL, 250 μM) stored at -80°C in a 1/1 v/v water / glycerol mixture was diluted with buffer (130 μL) and with a freshly-prepared DTT buffer solution (10 μL, 10 mM) and incubated for 1 h at 4°C to dissociate protein dimers. The solution was subsequently washed with buffer (3×500 μL) in a 5 kDa centricon. Afterwards, the sample was concentrated (to 50 μL) and DyIII-maleimido complex (DOTA or DTPA) buffer solution (0.7 μL, 26.7 mM, 3 equiv.) and additional amount of buffer (150 μL) were added. The solution was incubated for 1 h at r.t. and then washed with buffer (3×500 μL) in a 5 kDa centricon and concentrated to 100 μL. K1 spin concentration was checked by CW EPR. Samples were further concentrated (to 25 μL), glycerol (25 μL) was added, and the samples were filled into EPR quartz tubes of outer diameter of 2.95 mm. As the presence of dissolved oxygen may affect RE measurements, dry nitrogen was blown over each sample before immersion of the sample tube into liquid nitrogen. It was previously shown that such treatment significantly reduces the effect of paramagnetic oxygen on the DyIII-induced RE.[60] After preparation, samples were stored in liquid nitrogen.
To provide a diamagnetic reference, protein samples labeled with [Lu(DOTA)] were prepared for each T4-lysozyme mutant analogous to the described DyIII-labeling procedure. As the longitudinal relaxation of nitroxide radicals in the absence of DyIII is mainly determined by their local environment, the use of [Lu(DOTA)] label as a diamagnetic reference is feasible for both [Dy(DOTA)]-and [Dy(DTPA)]−- labeled samples.
Supplementary Material
Acknowledgments
We acknowledge financial support by the SNF (Grant No. 200021_121579), NIH grant EY05216, and the Jules Stein Professorship (to W.L.H.). We are grateful to Professors Kalman Hideg and Tamas Kalai for the generous gift of the hydroxylamine spin label used in these experiments.
References
- 1.Brockman J, Nelson B, Corn R. Annu Rev Phys Chem. 2000;51:41–63. doi: 10.1146/annurev.physchem.51.1.41. [DOI] [PubMed] [Google Scholar]
- 2.Willets KA, Van Duyne RP. Annu Rev Phys Chem. 2007;58:267–297. doi: 10.1146/annurev.physchem.58.032806.104607. [DOI] [PubMed] [Google Scholar]
- 3.Weiss S. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 4.Giepmans B, Adams S, Ellisman M, Tsien R. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 5.Stryer L. Annu Rev Biochem. 1978;47:819–846. doi: 10.1146/annurev.bi.47.070178.004131. [DOI] [PubMed] [Google Scholar]
- 6.Selvin P. Nat Struct Mol Biol. 2000;7:730–734. doi: 10.1038/78948. [DOI] [PubMed] [Google Scholar]
- 7.Milov A, Salikhov K, Shchirov M. Fiz Tverd Tela (Leningrad) 1981;23:975–982. [Google Scholar]
- 8.Milov A, Ponomarev A, Tsvetkov Y. Chem Phys Lett. 1984;110:67–72. [Google Scholar]
- 9.Martin R, Pannier M, Diederich F, Gramlich V, Hubrich M, Spiess H. Angew Chem Int Ed. 1998;37:2834–2837. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2833::AID-ANIE2833>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 10.Pannier M, Veit S, Godt A, Jeschke G, Spiess H. J Magn Reson. 2000;142:331340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 11.Jeschke G. Chem Phys Chem. 2002;3:927–932. doi: 10.1002/1439-7641(20021115)3:11<927::AID-CPHC927>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 12.Fajer P. J Phys: Condens Matter. 2005;17:1459–1469. [Google Scholar]
- 13.Borbat PP, Freed JH. Meth Enzymol. 2007;423:52–116. doi: 10.1016/S0076-6879(07)23003-4. [DOI] [PubMed] [Google Scholar]
- 14.Bhatnagar J, Freed J, Crane B. Meth Enzymol. 2007;423:117–133. doi: 10.1016/S0076-6879(07)23004-6. [DOI] [PubMed] [Google Scholar]
- 15.Jeschke G, Polyhach Y. Phys Chem Chem Phys. 2007;9:1895–1910. doi: 10.1039/b614920k. [DOI] [PubMed] [Google Scholar]
- 16.Schiemann O, Prisner T. Q Rev Biophys. 2007;40:1–53. doi: 10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- 17.Tsvetkov YD, Milov AD, Maryasov AG. Russ Chem Rev. 2008;77:487–520. [Google Scholar]
- 18.Klare JP, Steinhoff HJ. Photosynth Res. 2009;102:377–390. doi: 10.1007/s11120-009-9490-7. [DOI] [PubMed] [Google Scholar]
- 19.Jeschke G. Annu Rev Phys Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 20.Bordignon E. Topics Curr Chem. 2012;321:121–157. doi: 10.1007/128_2011_243. [DOI] [PubMed] [Google Scholar]
- 21.Krstic I, Endeward B, Margraf D, Marko A, Prisner T. Topics Curr Chem. 2012;321:159–198. doi: 10.1007/128_2011_300. [DOI] [PubMed] [Google Scholar]
- 22.Phan G, Remaut H, Wang T, Allen WJ, Pirker KF, Lebedev A, Henderson NS, Geibel S, Volkan E, Yan J, Kunze MBA, Pinkner JS, Ford B, Kay CWM, Li H, Hultgren SJ, Thanassi DG, Waksman G. Nature. 2011;474:49–53. doi: 10.1038/nature10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, Tyl M, Ward R, Sobott F, Maman J, Murthy AS, Watson AA, Fedorov O, Bowman A, Owen-Hughes T, El Mkami H, Murzina NV, Norman DG, Laue ED. Nat Struct Mol Biol. 2013;20:29–35. doi: 10.1038/nsmb.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Georgieva ER, Borbat PP, Ginter C, Freed JH, Boudker O. Nat Struct Mol Biol. 2013;20:215–221. doi: 10.1038/nsmb.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haenelt I, Wunnicke D, Bordignon E, Steinhoff HJ, Slotboom DJ. Nat Struct Mol Biol. 2013;20:210–214. doi: 10.1038/nsmb.2471. [DOI] [PubMed] [Google Scholar]
- 26.Todd A, Cong J, Levinthal F, Levinthal C, Hubbell W. Proteins. 1989;6:294–305. doi: 10.1002/prot.340060312. [DOI] [PubMed] [Google Scholar]
- 27.Hubbell W, Gross A, Langen R, Lietzow M. Curr Opin Struct Biol. 1998;8:649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- 28.Hubbell W, Cafiso D, Altenbach C. Nat Struct Mol Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 29.Narr E, Godt A, Jeschke G. Angew Chem Int Ed. 2002;41:3907–3910. doi: 10.1002/1521-3773(20021018)41:20<3907::AID-ANIE3907>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 30.Jeschke G, Zimmermann H, Godt A. J Magn Reson. 2006;180:137–146. doi: 10.1016/j.jmr.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 31.Bode B, Plackmeyer J, Prisner T, Schiemann O. J Phys Chem A. 2008;112:5064–5073. doi: 10.1021/jp710504k. [DOI] [PubMed] [Google Scholar]
- 32.Lueders P, Jeschke G, Yulikov M. J Phys Chem Lett. 2011;2:604–609. doi: 10.1021/jz300316q. [DOI] [PubMed] [Google Scholar]
- 33.Reginsson GW, Kunjir NC, Sigurdsson ST, Schiemann O. Chem Eur J. 2012;18:13580–13584. doi: 10.1002/chem.201203014. [DOI] [PubMed] [Google Scholar]
- 34.Yulikov M, Lueders P, Farooq Warsi M, Chechik V, Jeschke G. Phys Chem Chem Phys. 2012;14:10732–10746. doi: 10.1039/c2cp40282c. [DOI] [PubMed] [Google Scholar]
- 35.Kaminker I, Yagi H, Huber T, Feintuch A, Otting G, Goldfarb D. Phys Chem Chem Phys. 2012;14:4355–4358. doi: 10.1039/c2cp40219j. [DOI] [PubMed] [Google Scholar]
- 36.Lueders P, Jäger H, Hemminga MA, Jeschke G, Yulikov M. J Phys Chem B. 2013;117:2061–2068. doi: 10.1021/jp311287t. [DOI] [PubMed] [Google Scholar]
- 37.Kulikov A, Likhtenstein G. Adv Mol Relax Inter Proc. 1977;10:47–79. [Google Scholar]
- 38.Jäager H, Koch A, Maus V, Spiess H, Jeschke G. J Magn Reson. 2008;194:254–263. doi: 10.1016/j.jmr.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 39.Lueders P, Razzaghi S, Jäager H, Tschaggelar R, Hemminga M, Yu-likov M, Jeschke G. Mol Phys. 2013 in press. [Google Scholar]
- 40.Eaton S, Eaton G. Biol Magn Reson. 2000;19:347–381. [Google Scholar]
- 41.Zhou Y, Bowler B, Lynch K, Eaton S, Eaton G. Biophys J. 2000;79:1039–1052. doi: 10.1016/S0006-3495(00)76358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ulyanov D, Bowler BE, Eaton GR, Eaton SS. Biophys J. 2008;95:5306–5316. doi: 10.1529/biophysj.108.141887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato H, Kathirvelu V, Spagnol G, Rajca S, Rajca A, Eaton SS, Eaton GR. J Phys Chem B. 2008;112:2818–2828. doi: 10.1021/jp073600u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fielding AJ, Usselman RJ, Watmough N, Slinkovic M, Frerman FE, Eaton GR, Eaton SS. J Magn Reson. 2008;190:222–232. doi: 10.1016/j.jmr.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allegrozzi M, Bertini I, Janik M, Lee Y, Lin G, Luchinat C. J Am Chem Soc. 2000;122:4154–4161. [Google Scholar]
- 46.Bertini I, Lee Y, Luchinat C, Piccioli M, Poggi L. ChemBioChem. 2001;2:550–558. doi: 10.1002/1439-7633(20010803)2:7/8<550::AID-CBIC550>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 47.Bertini I, Donaire A, Jimenez B, Luchinat C, Parigi G, Piccioli M, Poggi L. J Biomol NMR. 2001;21:85–98. doi: 10.1023/a:1012422402545. [DOI] [PubMed] [Google Scholar]
- 48.Bertini I, Janik M, Lee Y, Luchinat C, Rosato A. J Am Chem Soc. 2001;123:4181–4188. doi: 10.1021/ja0028626. [DOI] [PubMed] [Google Scholar]
- 49.Barbieri R, Bertini I, Cavallaro G, Lee Y, Luchinat C, Rosato A. J Am Chem Soc. 2002;124:5581–5587. doi: 10.1021/ja025528d. [DOI] [PubMed] [Google Scholar]
- 50.Su XC, Otting G. J Biomol NMR. 2010;46:101–112. doi: 10.1007/s10858-009-9331-1. [DOI] [PubMed] [Google Scholar]
- 51.Keizers PM, Ubbink M. Prog Nucl Magn Reson Spectrosc. 2011;58:88–96. doi: 10.1016/j.pnmrs.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 52.Voss J, Wu J, Hubbell W, Jacques V, Meares CF, Kaback H. Biochemistry. 2001;40:3184–3188. doi: 10.1021/bi002333e. [DOI] [PubMed] [Google Scholar]
- 53.Fleissner M, Brustad E, Kalai T, Altenbach C, Cascio D, Peters F, Hideg K, Peuker S, Schultz P, Hubbell W. Proc Natl Acad Sci USA. 2009;106:21637–21642. doi: 10.1073/pnas.0912009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garbuio L, Bordignon E, Brooks E, Hubbell W, Jeschke G, Yulikov M. J Phys Chem B. 2013;117:3145–3153. doi: 10.1021/jp401806g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cotton S. Lanthanide and Actinide Chemistry. John Wiley & Sons, Ltd; 2006. [Google Scholar]
- 56.Lyubenova S, Siddiqui MK, Penning de Vries MJM, Ludwig B, Prisner TF. J Phys Chem B. 2007;111:3839–3846. doi: 10.1021/jp065805t. [DOI] [PubMed] [Google Scholar]
- 57.Blum H, Cusanovich M, Sweeney W, Ohnishi T. J Biol Chem. 1981;256:2199–2206. [PubMed] [Google Scholar]
- 58.Ward R, Bowman A, Sozudogru E, El-Mkami H, Owen-Hughes T, Norman D. J Magn Reson. 2010;207:164–7. doi: 10.1016/j.jmr.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zagdoun A, Casano G, Ouari O, Lapadula G, Rossini AJ, Lelli M, Baffert M, Gajan D, Veyre L, Maas WE, Rosay M, Weber RT, Thieuleux C, Coperet C, Lesage A, Tordo P, Emsley L. J Am Chem Soc. 2012;134:2284–2291. doi: 10.1021/ja210177v. [DOI] [PubMed] [Google Scholar]
- 60.Lueders P, Jaeger H, Hemminga MA, Jeschke G, Yulikov M. J Phys Chem Lett. 2012;3:1336–1340. doi: 10.1021/jz300316q. [DOI] [PubMed] [Google Scholar]
- 61.Doll A, Pribitzer S, Tschaggelar R, Jeschke G. J Magn Reson. 2013;230:27. doi: 10.1016/j.jmr.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 62.Barge A, Cravotto G, Gianolio E, Fedeli F. Contrast Media Mol Imaging. 2006;1:184–188. doi: 10.1002/cmmi.110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.