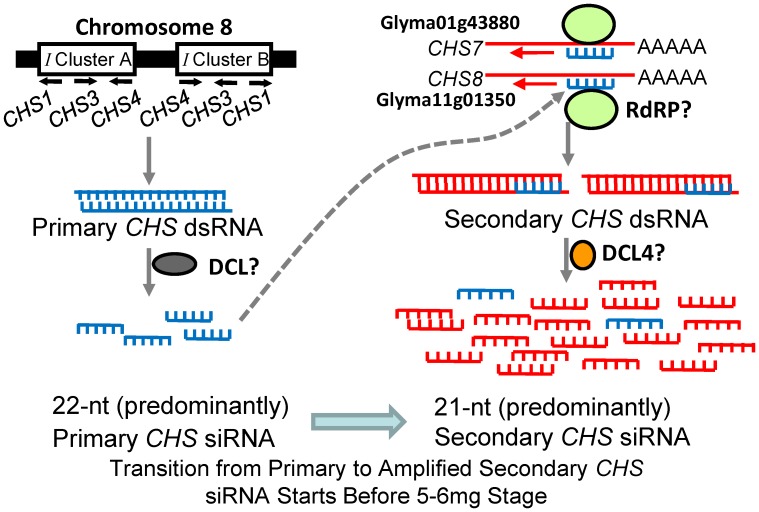
Figure 5. Biogenesis of CHS siRNAs Preventing Pigment Formation in Developing Soybean Seed Coats of the Williams Cultivar with ii Genotype.
Partially adapted from [4]. The long inverted repeat of the ii allele of the I locus on Chromosome 8 contains cluster A (CHS1-3-4) and cluster B (CHS4-3-1) and forms the nascent CHS dsRNA although the exact size is unknown. The cleavage of CHS dsRNA by putative Dicer Like proteins (DCL) generates predominantly 22-nt primary CHS siRNAs at the earliest developmental stages, resulting in the degradation in trans of targeted transcripts from CHS7 (Glyma01g43880) and CHS8 (Glyma11g01350) located on Chromosomes 1 and 11, respectively. After cleavage at the mRNA sites targeted by the primary CHS siRNAs, a putative RdRP synthesizes dsRNA from the cleaved CHS mRNA. The 21-nt secondary CHS siRNAs likely generated by DCL4 homologs from this dsRNA can target additional CHS mRNAs, thus amplifying the silencing response as well as spreading it over a larger region of the target. The transition from predominantly 22-nt CHS siRNAs (shown in blue) representing the origin of the CHS1-3-4 siRNAs to the 21-nt secondary CHS siRNAs (shown in red) representing the target CHS7 and CHS8 mRNAs occurs at the 5–6 mg seed stage as indicated.