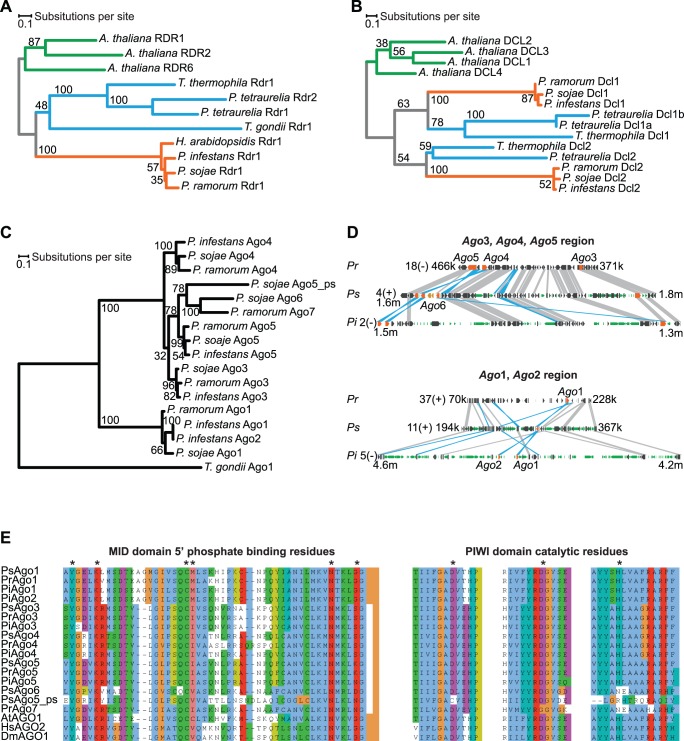
Figure 1. RNA silencing machinery in Phytophthora.
(A–C) Phylogenetic analysis of Rdr, Dcl and Ago proteins, respectively. Protein domains used for analysis were the Rdr RdRP domain, the Dcl RNaseIII A and B domains and the Ago DUF1785-PAZ-PIWI region. Maximum Likelihood bootstrap support values are shown. In (A) and (B), taxonomic groups are color-coded: viridiplantae (green), alveolates (blue) and oomycetes (orange). (D) Synteny maps of regions containing Ago3–5 (top) and Ago1/2 (bottom) in P. ramorum (Pr), P. sojae (Ps), and P. infestans (Pi). Regions are labeled with scaffold numbers, alignment orientation (+/−) and coordinate ranges. Genes (dark arrows), Ago genes (orange arrows), and transposable elements (green arrows) are depicted. Conserved intervals are connected by gray (direct) or blue (inverse) boxes (E) MAFFT alignment with ClustalX coloring of amino acid residues from the Ago MID (5′ phosphate binding) and PIWI (catalytic) domains. Positions shown to interact with the guide-strand 5′ monophosphate (MID), and the Mg2+-coordinating catalytic residues (PIWI) are marked with an asterisk [40], [41], [42], [43], [44], [45], [46].