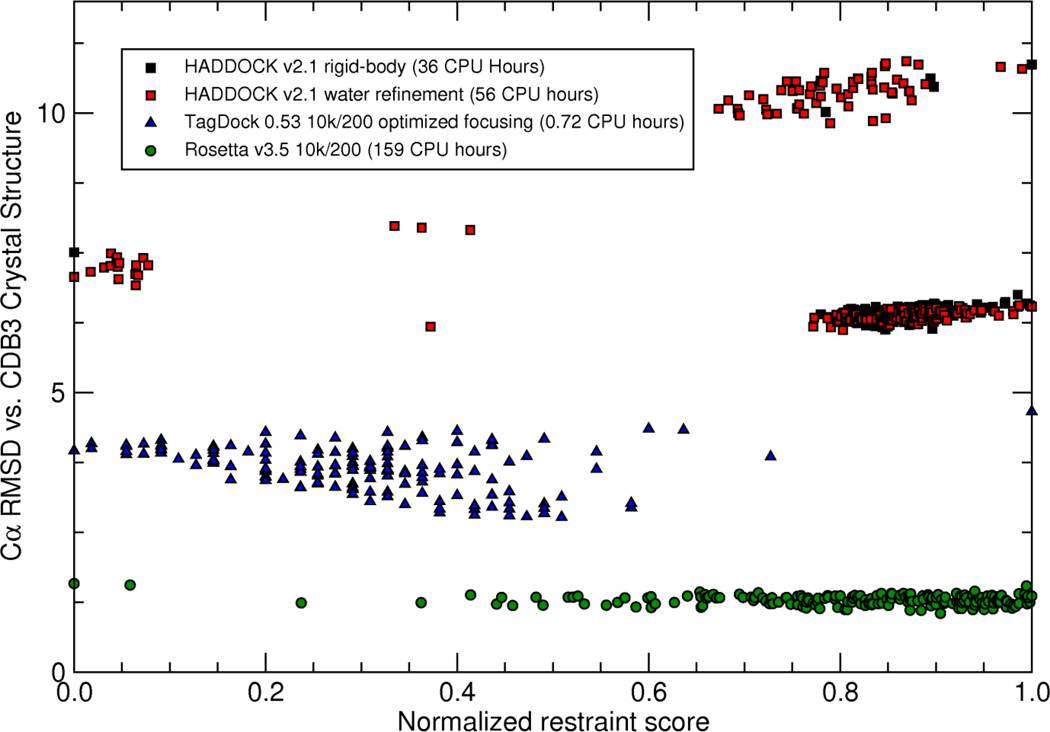
Figure 3. CDB3 Homodimer Docking from 18 experimentally measured (DEER) distance restraints using HADDOCK 2.1, RosettaDock 3.5, and TagDock.
10,000 decoys were computed with each program, and the 200 models that best satisfy the experimental restraints were selected for analysis. RosettaDock (green circles) produces atomic-resolution structures with this restraint set, with the best model just 1.1Å Cα RMSD from chains P and Q of the 1HYN crystal structure. TagDock (blue triangles) produces intermediate-resolution structures with Cα RMSD as small as 2.8Å from the crystal structure. HADDOCK (red and black squares) had trouble assembling the interdigitating dimer arms of CDB3 (best Cα RMSD of 6.1Å). We suspect this is due to a more limited search of pose space during its rigid-body phase (black squares), which we observed to produce a large number of degenerate structures. For example, the best-scoring rigid-body HADDOCK pose depicted at the far left of the graph was replicated 16 times in the 10,000 output poses. These degenerate structures resolved into unique structures only after the high-resolution refinement stage (red squares).