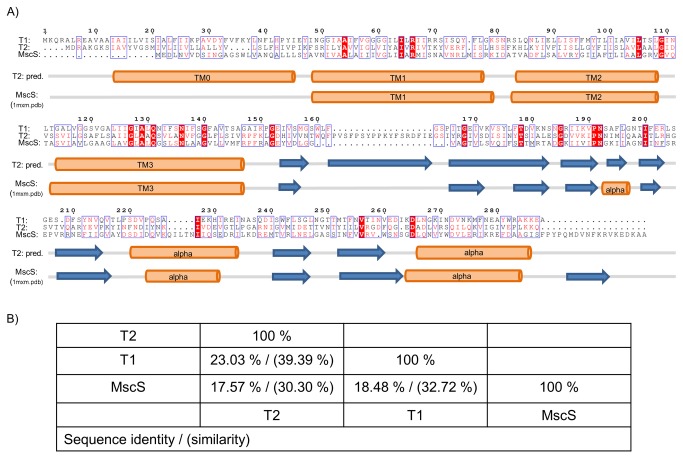
Figure 1. Sequence alignment and secondary structure of bacterial and archeal mechanosensitive channels.
(A) Sequence comparison of the archeal mechanosensitive channels T1 and T2 from Thermoplasma volcanium with MscS from E. coli. Identical and similar residues are boxed and color coded in red. Highest sequence similarity is found in transmembrane helix 3. Secondary structure elements based on the bioinformatic prediction tool Jpred 3 for T2 [65] and on the crystal structure of MscS (1mxm.pdb) is shown underneath the alignment. (B) Sequence identities and similarities between T1, T2 and MscS as calculated using the pairwise identity server SIAS [66].