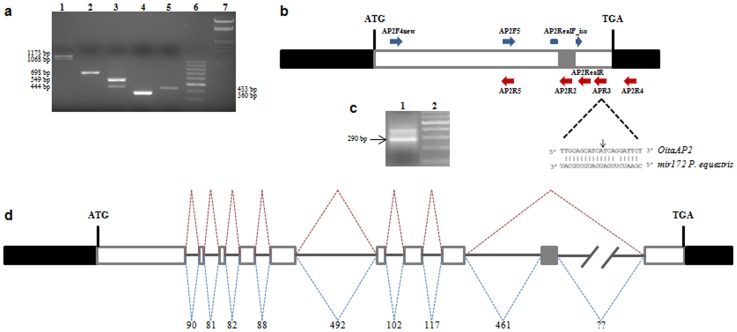
Figure 1. Alternative splicing, mir172 target site and genomic organization of the OitaAP2 gene of O. italica.
(a) Agarose gel electrophoresis of the PCR amplification products of the OitaAP2 cDNA with different primer pairs: lane 1, AP2F4new/AP2RealR; lane 2, AP2F4new/AP2R5; lane 3, AP2F5/APR3; lane 4, AP2RealF_iso/AP2R4; lane 5, AP2F5/AP2R2; lane 6, 100 bp DNA ladder (Thermo Scientific); lane 7, DNA Marker III (Thermo Scientific). (b) Schematic representation of the OitaAP2 cDNA with the relative positions of the primers used in the PCR amplifications. Black boxes represent the 5′- and 3′-UTRs; white boxes represent the exons; gray box represents the alternatively spliced region; dotted lines highlight the region corresponding to the miR172 target site with the black arrow indicating the cleavage point. (c) Agarose gel electrophoresis of the PCR amplification product of the OitaAP2 cDNA fragment cleaved by miR172 (lane 1); lane 2, 100 bp DNA ladder (Thermo Scientific). (d) Schematic representation of the genomic organization of the OitaAP2 gene. Black boxes represent the 5′- and 3′-UTRs; white boxes represent the exons; gray box represents the alternatively spliced exon; continuous lines represent introns; dotted lines indicate the two alternative splicing events; numbers indicate the length of introns (bp); question marks indicate the unknown size of intron 9.