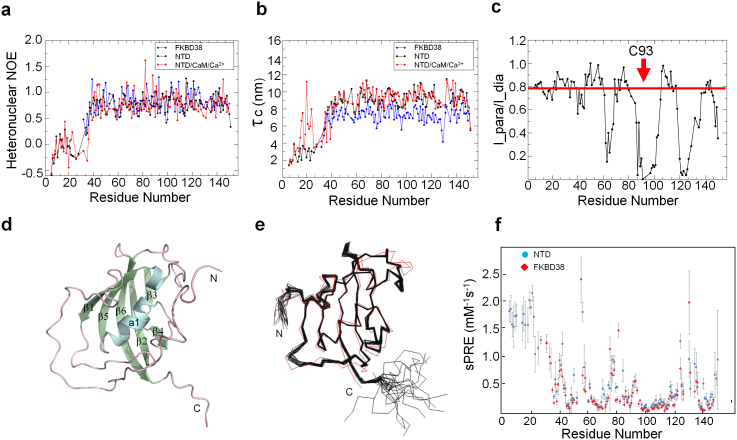
Figure 2. Relaxation, PRE data and NTD structure.
(a) {1H}-15N hetNOE for the backbone amide of the core isomerase domain (FKBD38; blue), N-terminal domain (NTD; black) of FKBP38, and the NTD/CaM/Ca2+ complex at 1:3 morlar ratio (red). The N-terminal tail (residues 1–32) exhibits negative or near zero {1H}-15N hetNOE values, suggesting the unstructured characteristic of this region. The estimation errors were not plotted for clarity but referable to Table 2. (b) The rotational correlation time τc was derived from the T1/T2 ratio for all residues. (c) Experimental signal intensity ratio I_para/I_dia from 2D 1H-15N HSQC spectra without and with the addition of twenty molar equivalents of ascorbic acid. Due to the slight protein concentration change after adding ascorbic acid, the intensity ratios were normalized by setting largest I_para/I_dia ratio to 1. A prosxyl spin label 3-(2-iodoacetamido)-proxyl (Sigma-Aldrich) was attached to residue Cys93 of NTD_C121A/C139T, indicated by the red arrow. The red line across the graph represented a threshold of 0.78, protons above which were considered far from the prosxyl spin label and unrestrained. (d) The model for the NMR structure of NTD is shown in ribbon representation and colored by secondary structure (α-helices in cyan, β-sheets in green, and loops in purple). The N-terminal extension seen in the NMR structure was modeled from the PRE data, as shown in Fig. 2c and Supplementary Fig. 3. The N- and C-termini are labeled, as are the secondary structural elements. (e) The backbone ensemble of 20 superimposed NMR-derived structures of the NTD (shown in black) and N-Cα-C′-trace of the isomerase core structure determined crystallographically (PDB ID 2AWG, shown in red). The N-terminal extension (residues 1–32) is not presented for clarity as it is highly flexible and only a restricted number of NOEs were observed in this region. Molecular graphics were created using PyMOL (http://www.pymol.org). (f) Solvent PRE experiments for NTD and FKBD38.