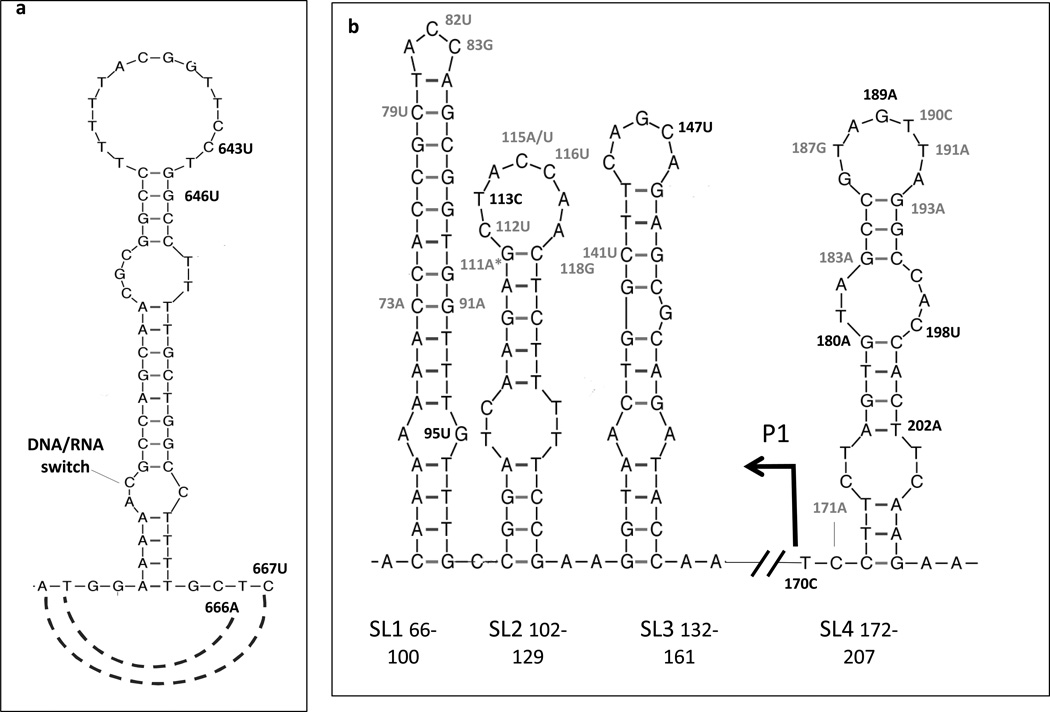
Fig. 1. Location of plasmid dosage-modulating mutations within known plasmid copy number regulatory sites.
Methylating agent-selected ALKBH2 mutations located in sites controlling plasmid replication initiation are shown in black. For comparison, previously-described mutations increasing ColE1 plasmid copy number are shown in grey (their references in the literature can be found in (3)). Graphic representations were generated using the mfold program (22), which predicts secondary structures of single-stranded nucleic acids based on a thermodynamic model. a. Area surrounding the DNA/RNA switch, where R-loop formation, primer processing and Pol I extension occur. b Antisense RNA I regulatory elements (SL 1, 2, 3 and promoter P1) and pre-primer SL4.