Figure 1.
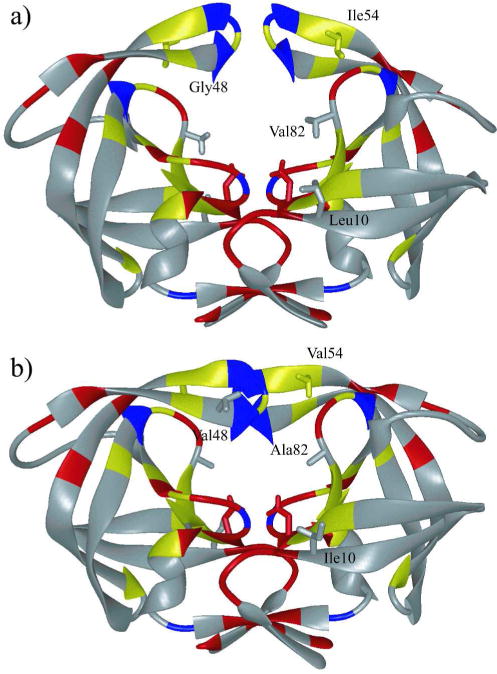
HIV-1 protease colored by sequence conservation (41). Residues colored red are invariant in both untreated and treated populations of patients. Residues colored yellow are invariant only in the untreated population. Invariant glycine residues are colored blue. (a) The protease structure in unliganded (PDB code: 1HHP) and (b) liganded (PDB code: 1HXB) state. The side chains of the catalytic aspartic acids Asp25 in both monomers, as well as mutation sites Leu10, Ile54, and Val82 are displayed.
