Figure 3.
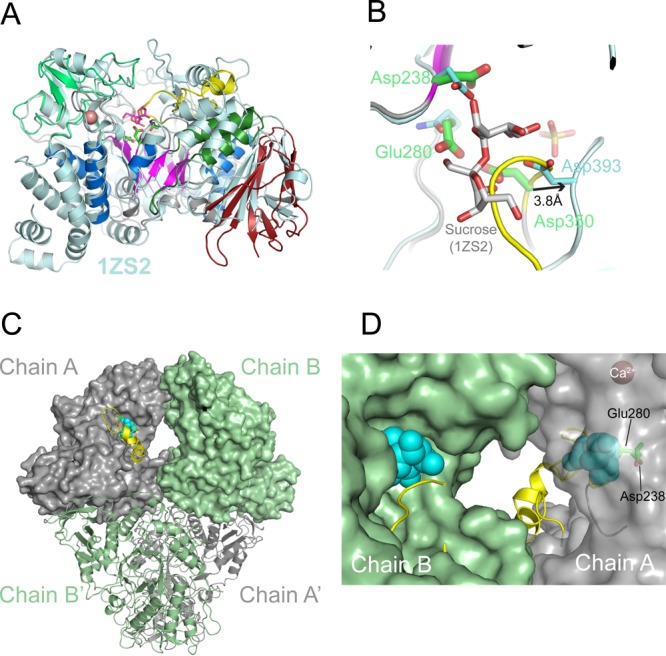
Structural homology and active site geometry of TreS. (A) Superposition of the TreS monomer with its closest structural neighbor, sucrose-bound structure of Neisseria polysaccharea amylosucrase (PDB entry 1ZS2(14)). Sticks in magenta indicate the amylosucrase-bound sucrose, and sticks in green indicate the TreS catalytic residues (Asp238, Glu280, and Asp350), with Ca2+ site as a sphere in salmon. (B) Position of sucrose (sticks in gray) in the active site of TreS (chain A), based on the structural superposition in panel A. The side chains of the catalytic triad of TreS (green) and amylosucrase (cyan) are shown. The β7−β8 loop (yellow) overlaps with the putative substrate binding site. (C) Molecular surfaces of chains A and B in the TreS tetramer. Spheres in light blue represent sucrose according to the superposition in panel A, while the β7−β8 loop is shown as a ribbon (yellow). (D) Close-up view of the cavity between the active sites of chains B and A (view of panel C rotated by 180° about the vertical axis). The surface of chain A is left transparent to reveal the location of the catalytic residues and of the (modeled) substrate.
