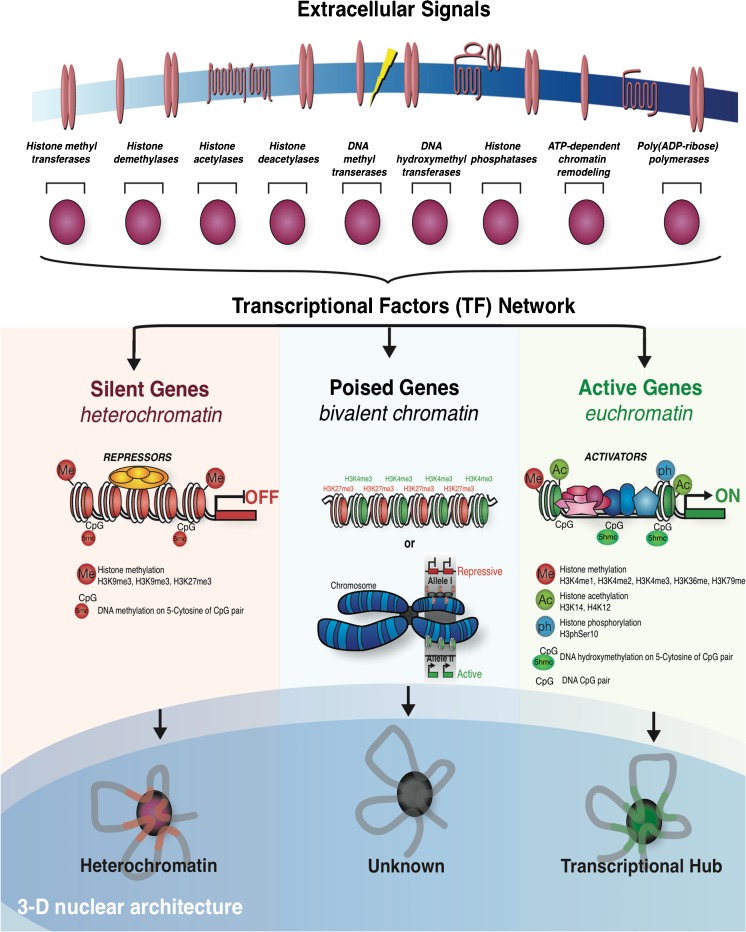
Fig. 1.
Dynamic genome architecture in the nuclear space: regulation of gene expression in 3 dimensions. Epigenetic response to extrinsic signals occurs through the transcriptional factors network. Chromosomes are organized into loose (euchromatin) and highly condensed (heterochromatin) domains. Heterochromatin is transcriptionally repressive, whereas euchromatin is transcriptionally permissive. Bivalent chromatin (middle) that poise genes for transcriptional activation are thought to be either superposition of active and silent epigenetic marks, or simply represent monoallelic gene expression. An 11-nm “beads-on-a-string” chromatin fiber is comprised of nucleosomal arrays and has an additional level of organization in the context of 3-dimensional (3D) nuclear volume (bottom). The high level of transcriptional regulation depends on the 3D organization and functional compartmentalization of the nucleus [42, 43]. In mammalian cells, transcription of genes that are concomitantly activated in response to extracellular stimuli or during cell differentiation occurs specifically at intranuclear foci enriched with active RNA polymerase II (RNAPII) [142]. These transcriptional hubs are known as transcription factories and, in addition to RNAPII, they often include transcription factors (TF) [41]. The distribution of DNA methylation and a small subset of post-translational histone marks represents different flavors of chromatin related either to gene activation (right) or gene repression (left). ATP adenosine triphosphate; ADP adenosine diphosphate; Me methylation; 5 m 5-methylcytosine; Ac acetylation; 5hm 5-hydroxymethylcytosine; ph phosphorylation