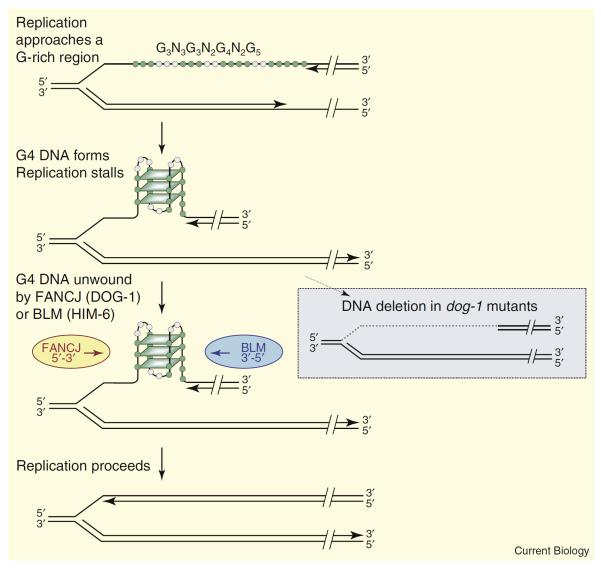
Figure 1.
G4 DNA formation and repair at a replication fork.
The figure shows G4 DNA formed within the sequence G3N3G3N2G4N2G5. G, green; other bases, gray; parallelograms represent G-quartets. Structure formation is illustrated in the lagging strand, which is thought to be more prone to formation of alternative structures because it persists in a denatured state longer than the leading strand. Only a single structure is shown in the figure, but note that three of the guanines in the sequence that are shown outside of the quartets could be included by changing the register of individual G-runs, and thus contribute to structural polymorphism. FANCJ (C. elegans DOG-1) or BLM (C. elegans HIM-6) unwind G4 DNA to allow replication to proceed (left). In C. elegans dog-1 mutants, deletion occurs, with the 3′ boundary defined by the 3′ end of the G4 signature motif, and the 5′ boundary possibly determined by the location of the replication origin (right, dotted gray line indicates the deleted region).