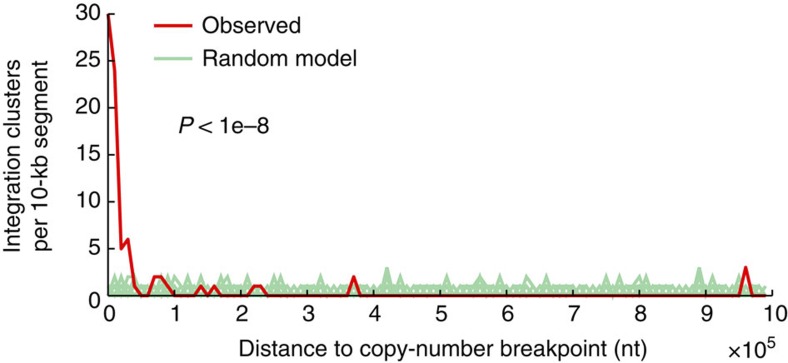
Figure 4. Integrations in HNSC colocalize with DNA copy-number breakpoints.
One hundred and ten HPV integration clusters (31 unique integrations) were compared with copy-number breakpoints determined using segmented Affymetrix SNP6 microarray data from TCGA. The distance to the nearest breakpoint was calculated for each cluster, and the observed distribution was tested for non-random colocalization by comparing with a uniform random integration model (P<1e−8 based on 1e−8 randomizations; 100 shown). Integration clusters (41.8%) were within 10 kb, whereas the random expectation was <0.5%. Ten kilobases are close to the SNP6 mapping resolution (average probeset spacing ~3 kb).