Figure 6.
Regulated high curvature vesicle tethering by the EAT domain
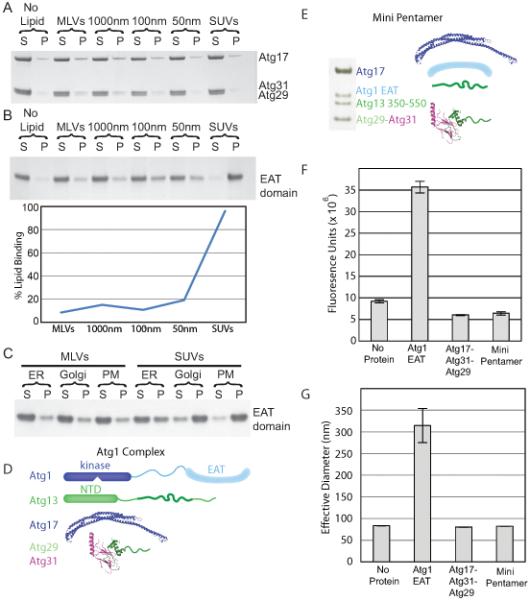
(A) Liposome sedimentation assays for the full-length Atg17-Atg31-Atg29 complex with Folch liposomes of varying diameters. Atg17-Atg31-Atg29 does not bind to Folch liposomes of any size.
(B) Liposome sedimentation assay for the EAT domain with Folch liposomes of varying diameters. Atg1 shows a strong preference for small sonicated Folch liposomes.
(C) Liposome sedimentation assay for the EAT domain with liposomes mimicking the endoplasmic reticulum (ER), golgi apparatus and plasma membrane (PM) of S. cerevisiae.
(D) SDS-PAGE of the minimal Atg1 complex. The bands for Atg1, 13, 17, 29 and 31 are labeled. Atg29 and Atg31 run at identical locations on SDS-PAGE.
(E) Schematic of the full-length Atg1 complex, for comparison to the mini-pentamer expressed in (D).
(F) Sonicated liposomes containing biotin were mixed with fluorescently labeled sonicated liposomes and buffer, the EAT domain, full-length Atg17-Atg31-Atg29 or the mini-pentamer. The EAT domain potently tethers vesicles, but Atg17-Atg31-Atg29 does not. Atg17-Atg31-Atg29 inhibits tethering by the EAT domain. Biotin liposomes were captured by streptavidin resin and the amount of fluorescent lipid tethered to the biotin liposomes was quantified.
(G) The effective diameter of SUVs obtained from light scattering with no protein, the EAT domain, full-length Atg17-Atg31-Atg29 or the mini-pentamer. The increase in the effective liposome diameter induced by the EAT domain shows that it tethers liposomes. The lack of increase above baseline shows that the other complexes tested do not tether liposomes, consistent with results in (F).
Error bars in (F) and (G) represent the standard deviation of triplicate experiments.
