Figure 1. Protocol for construction of Mu-seq libraries.
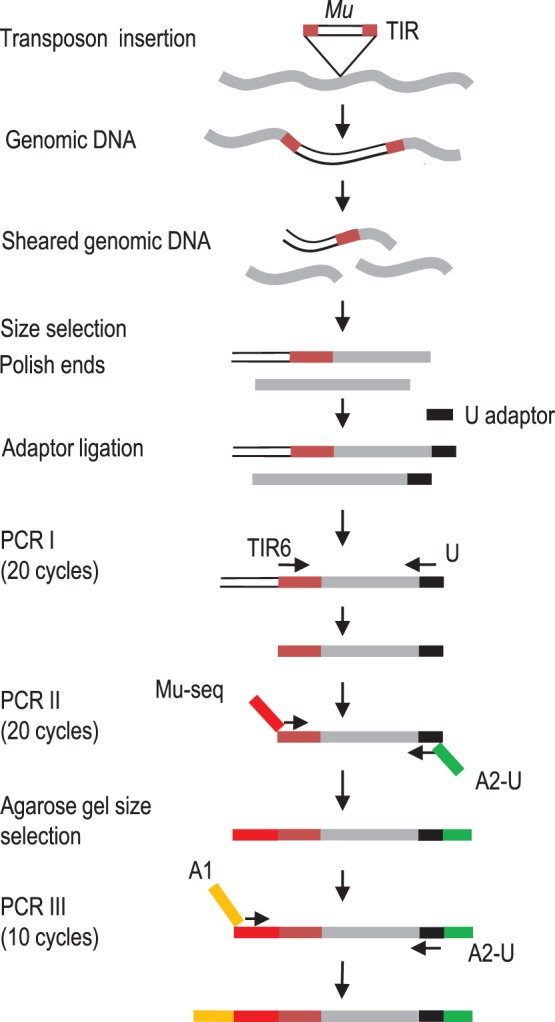
Genomic DNA from pools of 125 or more plants is sheared by sonication, size-enriched, made blunt-ended and ligated to an adaptor (U). The PCR I step uses a Mu-specific, TIR6 primer and a universal U adaptor primer to amplify genomic DNA fragments with TIRs at their 5′ ends. PCR II incorporates at the 5′-end, a Mu-seq primer that includes, from 5′ to 3′, part of the Illumina A1 sequencing adaptor, a 4-bp barcode, and a MuTIR region; and, at the 3′ end, an Ilumina, A2-U sequencing adaptor. PCR II products are size-selected by agarose gel electrophoresis. PCR III completes the nested synthesis of a complete A-1 Illumina sequencing adaptor. Colors highlight the TIR of a Mu transposon (brown), genomic DNA (grey), a universal adaptor U (black), the Mu-seq adaptor (red), an Illumina A2-U sequencing adaptor (green), and an Illumina A1 sequencing adaptor (orange).
