Figure 2. Structure of transposon-anchored Mu-seq reads.
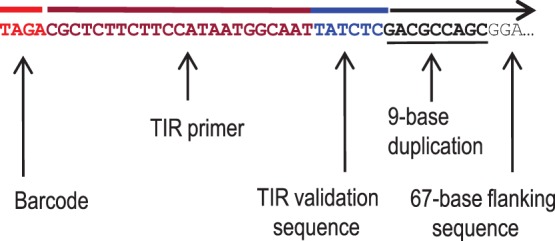
Each outward-directed, 100-base, Mu-seq sequence read includes a 27-base sequence derived from the Mu-seq adaptor [a 4-base, error-detecting, multiplex key (red), plus a 23-base, TIR-specific primer sequence (brown)]. The adjacent genomic sequence begins with 6 bases (TATCTC) from the Mu-TIR, 3′-end (blue) that are used to validate authentic Mu-flanking sequences (black). Validated reads are trimmed to remove the key and TIR sequences, then truncated if sequence quality at any point drops below a defined threshold. The remaining Mu-seq read (horizontal black arrow above sequence) thus includes one copy of the characteristic, 9-base, direct duplication created by the Mu transposon (underlined), and up to 67 bases of genomic sequence flanking the transposon insertion site (only 3 bases are shown). The barcode and library identifier are appended to the name of the trimmed read.
