Figure 4. Numbers of Mu-seq reads are reproducible in DNA from independent, pooled, tissue samples.
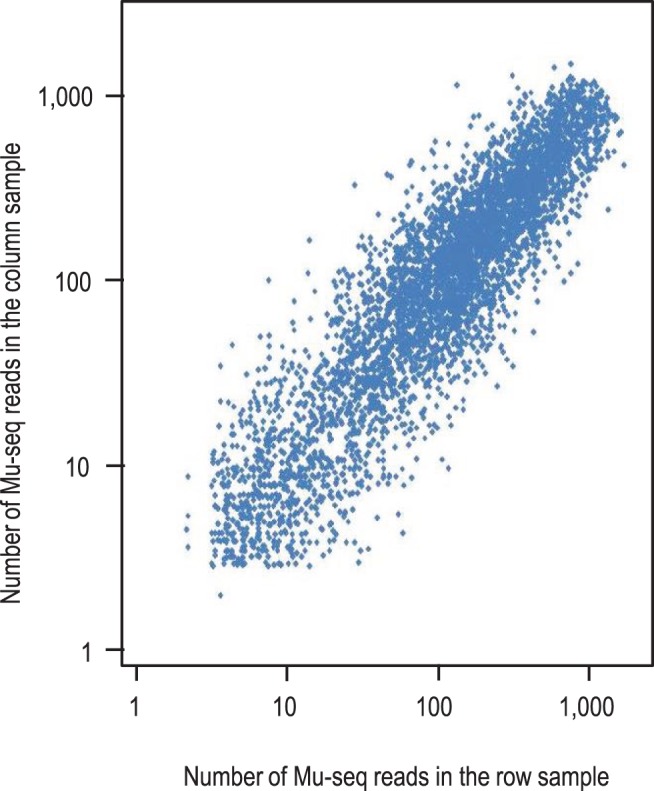
Novel transposon insertion sites were identified in a set of 576, Mu-inactive, maize families selected from the UniformMu population. The maize families were analyzed using a 48-sample, multiplex Mu-seq library sequenced in a single Illumina HiSeq II lane (see Table 1). Families represented by 2 to 4 plants were grown in a 24×24 grid array and DNA was prepared from pooled leaf samples of each row and column (24 families representing up to 96 plants per pool). Normalized numbers of Mu-seq reads are shown for row- and column- pools that yielded a total of 4,723 unique insertion sites. In 75% of instances where insertions were detected at points of axis intersection, Mu-seq read numbers differed by 2-fold or less. R2 = 0.72.
