Figure 6. Resolution of Mu insertions from different individuals where Mu targeted nearby positions in the genome.
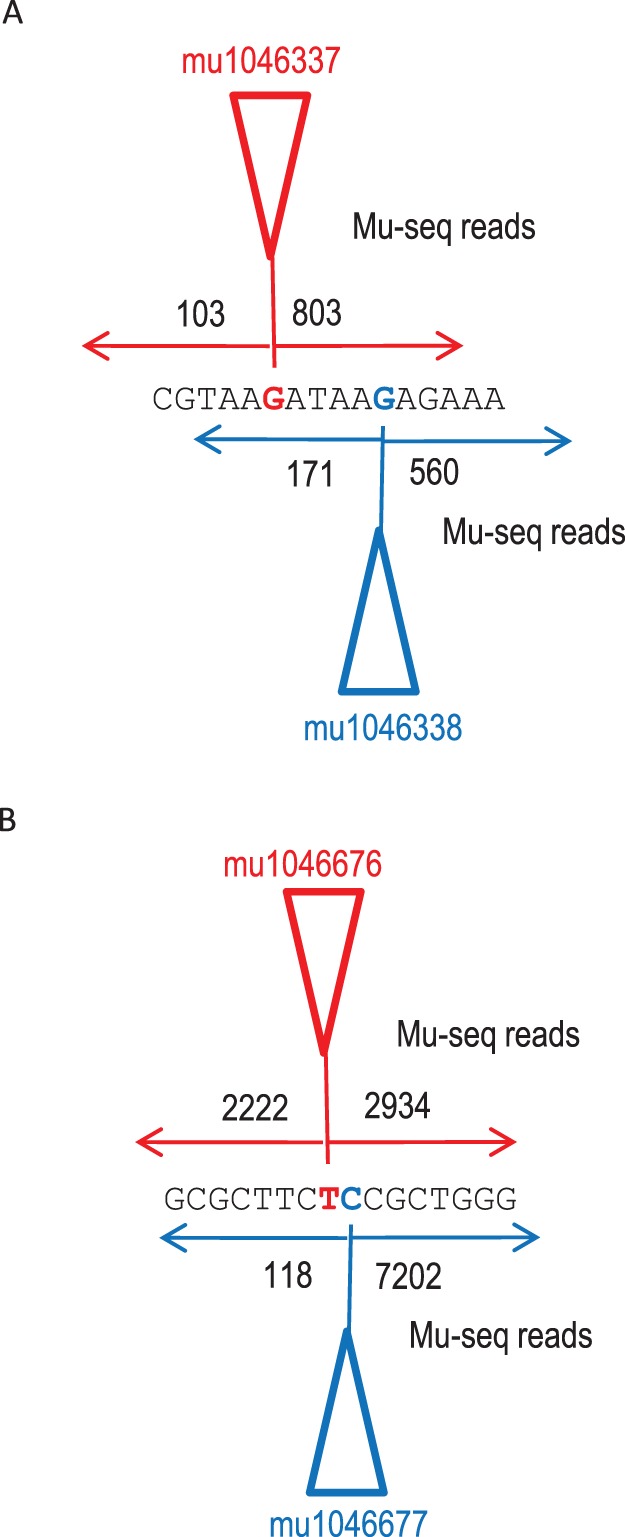
The two pairs of insertions shown in A and B were identified in a collection of 576 families analyzed in a single Mu-seq library. A ) A pair of insertion sites separated by 5 base-pairs on Chromosome 1. Sequence is shown between positions 19,615,632 and 19,615,648. Triangles depict insertion sites (red for insertion mu1046337) (blue for insertion mu1046338). B) A pair of insertions separated by a single base-pair on chromosome 1. Sequence is shown between positions 43,421,572 and 43,421,588. Triangles depict insertion sites (red for insertion mu1046676) (blue for insertion mu1046677). Numbers adjacent to each insertion site show the quantity of Mu-seq reads recovered from forward and reverse orientations. By convention, positions of Mu insertion sites are assigned to the left-most base of the 9-base, direct duplication, and in the orientation of the maize reference genome. Accordingly, start sites for alignments of sequence reads in the reverse orientation are shifted 8-bases to the left.
