Figure 8. Bioinformatic analysis of Mu-seq data.
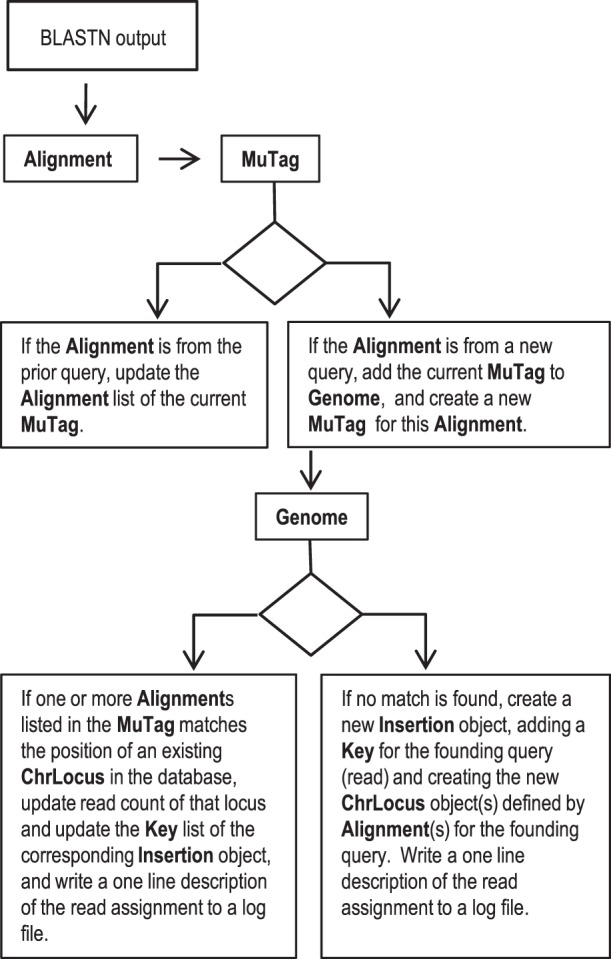
Trimmed Mu-seq reads from Illumina sequencing are aligned to the B73 maize reference genome using parallel BLASTN. Tab-delimited, BLAST output is parsed into Alignment objects. The top scoring Alignment(s) for each read are stored as MuTag objects. The Genome database assigns each MuTag to an Insertion object based on matching Alignment’s to the chromosome location(s) (ChrLocus objects) associated with each Insertion. Allowing insertions to have multiple loci accommodates ambiguity due to duplicate sequences in the maize genome.
