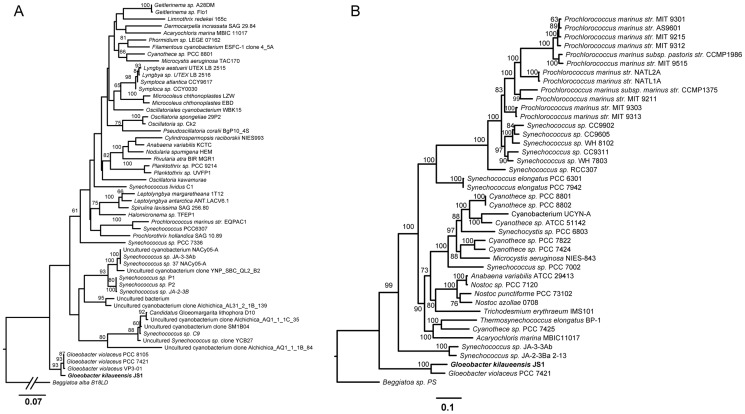
Figure 4. Maximum likelihood phylogenetic tree based on 16S rRNA gene sequences.
Sequences were aligned using Muscle, edited with Gblocks, and the tree was inferred using RAxML program with 100 bootstrap replicates and the GTR+Gamma model of rate substitution. The root of the tree was shortened to fit the figure (A). Sequences in this tree included the top 50 BLASTn matches of the Gloeobacter kilaueensis JS1T 16S rDNA sequence, among others. A phylogenetic tree was also constructed on the basis of 43 ribosomal proteins found in 41 cyanobacteria and the Beggiatoa sp. PS outgroup (B). Amino acid sequences of the 43 such proteins in each genome were individually aligned using Muscle, trimmed with Gblocks, and concatenated. Based on 5,359 aligned characters, the maximum likelihood analysis was performed using the RAxML program with the Gamma+WAG model of amino acid substitution and 100 bootstrap replicates, and visualized in FigTree.