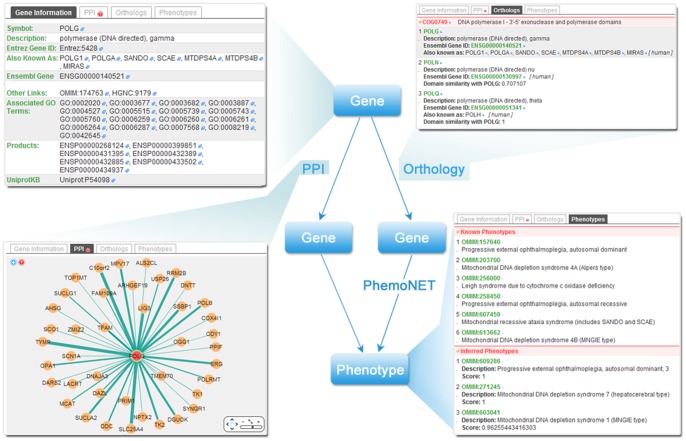
Figure 2. Main workflow and contents of the web Server.
Phenotypes of a gene are inferred via PPI paths and orthology paths. All elements in the web server are well annotated. Each element offers a cross-reference link that links to its original source or to the NCBI database. Genes are described by their names, descriptions, Entrez Gene ID, synonyms, Ensembl Gene IDs and other cross-reference links. Their products are identified by their Ensembl Protein IDs and UninprotKB Accessions. PPI information is presented by a network visualization tool, Cytoscape Web[61]. Orthologs are listed and grouped by the orthologous groups. The known phenotypes are listed above the inferred ones, which are sorted in a descendant order by the score.