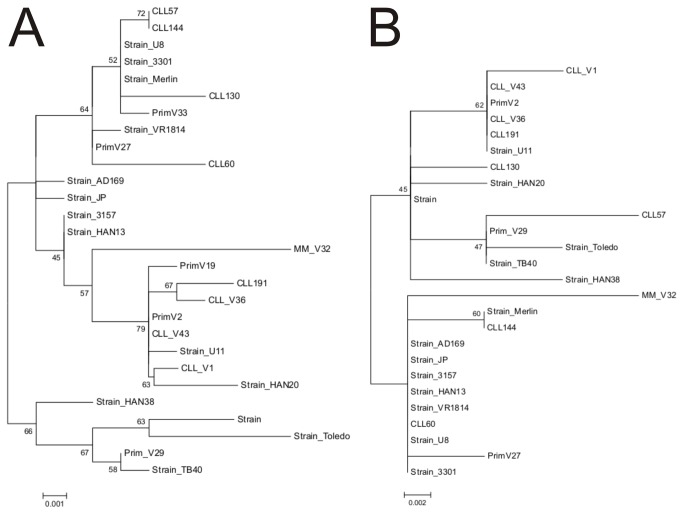
Figure 4. Phylogenetic analysis of the CMV UL32 gene.
(A) Analysis of the N-terminal aa50-784 of the UL32 gene from CMV strains detected in our study (CLL, n=7; multiple myeloma, n=1; immunocompetent patients with primary CMV infections, n=5). (B) Analysis of the C-terminal aa493-1037 of the UL32 gene from the same CMV strains (CLL, n=7; multiple myeloma, n=1; immunocompetent patients with primary CMV infections, n=3). Sequences were compared to the published sequences of 3 laboratory adapted CMV strains and 11 previously described clinical isolates (see methods section) using the Jones-Taylor-Thornton model for constructing the tree with the maximum likelihood algorithm in MEGA. Robustness of the nodes was assessed with the Kimura two-parameter model for neighbor-joining algorithms and bootstrap-resampling. Bootstrap values (% after 1000 iterations) are shown for major branches.