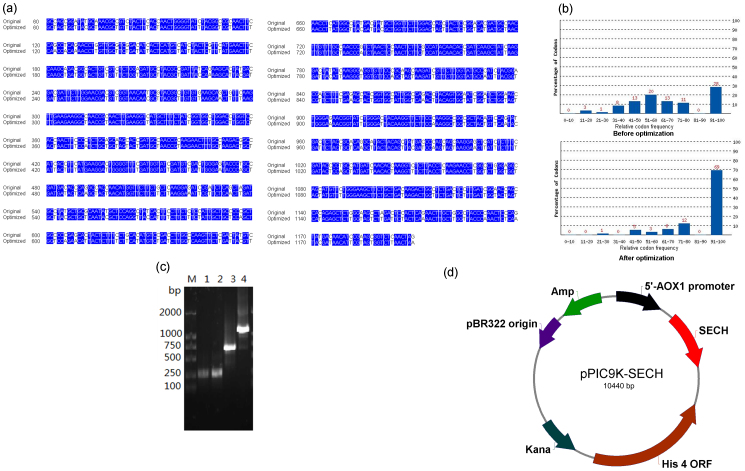
Figure 1.
(a) Alignment of codon-optimised and wild-type endochitinase cDNAs. Bases that are the same between codon-optimised and wild-type cDNAs are marked in blue. (b) Enhanced codon usage of endochitinase for expression in Pichia. (c) Successive PCR of the codon-optimised endochitinase cDNA. Lane M: DL2000 marker. Lanes 1–4: Assembled PCR products of the first group of primers (F1, R1, F2, R2, F3 and R3), the second group of primers (F12, R12, F13, R13, F14 and R14), the third group of primers (F4–F11 and R4–R11) and the above three DNA fragments amplified by primer set F1 + R1, respectively. (d) Schematic map of the constructed expression vector pPIC9K-SECH.