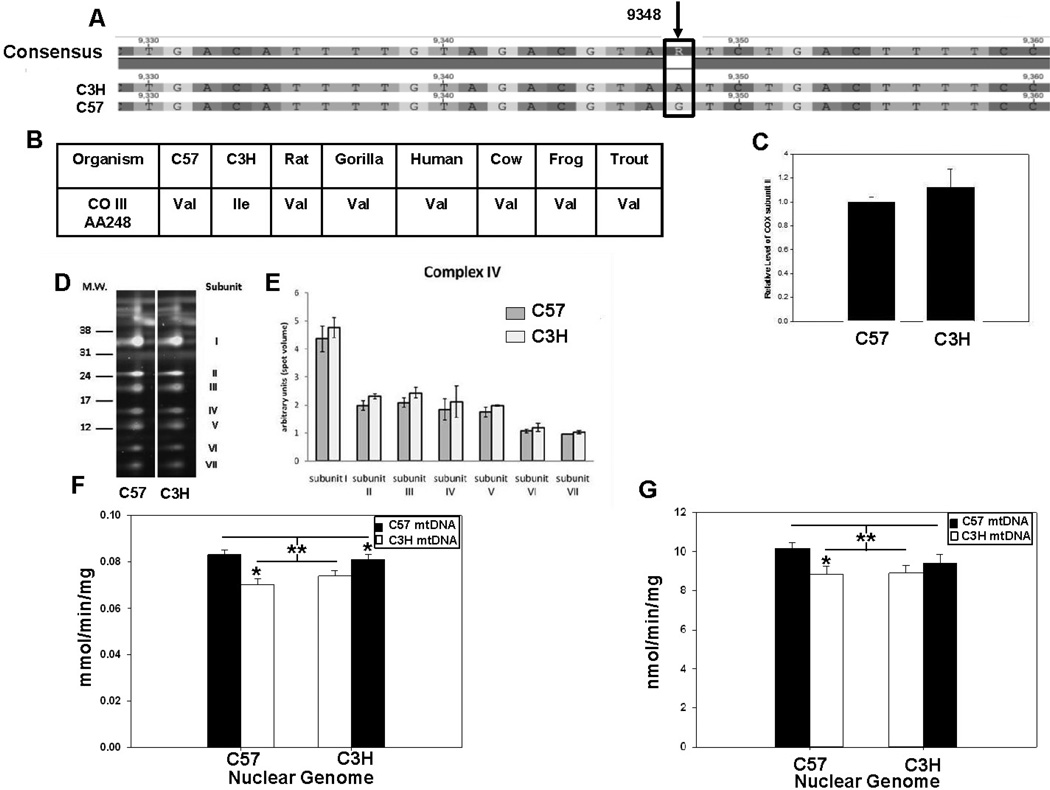
Figure 4. Complex IV (cytochrome oxidase) protein levels and activity in C57 and C3H mitochondria.
Proteomic and enzymatic analyses of heart mitochondrial isolates for cytochrome c oxidase. (A) MtDNA sequence of C3H and C57 mice showing a G to A transitional missense mutation in subunit III of complex IV. (B) Conservation of valine at AA248 throughout vertebrates. (C) Levels of cytochrome oxidase (COX) subunit II and (D) blue native 2D proteomics of additional cytochrome oxidase subunits. (E) Quantification of subunits, indicating no differences in the expression of isolated complex IV subunits. (F) Cytochrome c oxidase activity following the oxidation of reduced cytochrome c at 550 nm in cardiac mitochondria from control (C57n:C57mt and C3Hn:C3Hmt) and MNX (C57n:C3Hmt and C3Hn:C57mt) mice. The nuclear genome for each group is indicated on the X-axis, whereas the filled or open bars indicate the presence of either the C57 mtDNA or C3H mtDNA, respectively. An asterisk indicates a significant difference (*p < 0.05) exists between the nuclear matched control and MNX animal; the double asterisk (**) with lines indicates a significant difference exists between groups having the C57 versus C3H mtDNA. (G) Cytochrome c oxidase oxygen consumption was determined in cardiac mitochondria using ascorbate + TMPD from control (C57n:C57mt and C3Hn:C3Hmt) and MNX (C57n:C3Hmt and C3Hn:C57mt) mice. The nuclear genome for each group is indicated on the X-axis, whereas the filled or open bars indicate the presence of either the C57 mtDNA or C3H mtDNA, respectively. An asterisk indicates a significant difference (*p < 0.05) exists between the nuclear matched control and MNX animal; the double asterisk with lines indicates a significant difference exists between groups having the C57 versus C3H mtDNA.