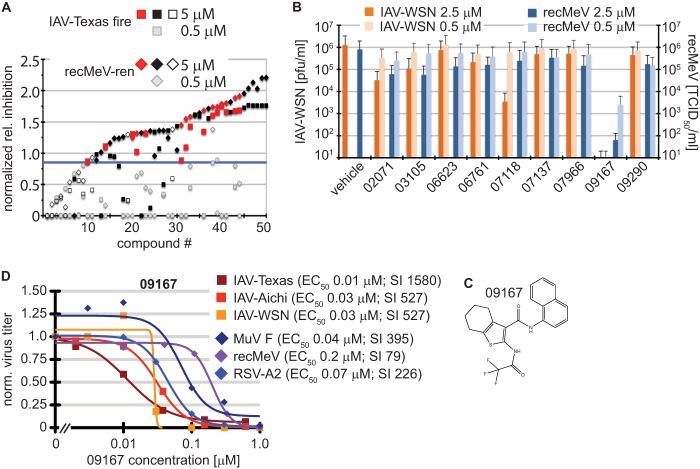
Fig 5.
Identification of a nanomolar pan-myxovirus inhibitor class. (A) Two-concentration confirmatory screen of the selected 51 hit candidates. Values were normalized to those for plate controls and represent inhibition relative to that of JMN3-003. Compounds are ordered by increasing relative inhibition values. Black filled symbols mark cytotoxic compounds (reducing cell metabolic activity by >25%), open symbols represent compounds that were inactive against at least one of the target viruses at 5 μM, and red symbols highlight the remaining hit candidates. Gray symbols show the corresponding normalized scores obtained for each entry at 0.5 μM. The blue line marks the hit cutoff (0.85) relative to JMN3-003 for all candidates at 5.0 μM. (B) Virus yield-based counterscreen of nine candidates at 2.5 and 0.5 μM against IAV-WSN and recMeV. Values represent averages for two independent experiments, and error bars show data ranges. (C) Chemical scaffold of lead candidate 09167 {2,2,2-trifluoro-N-[3-(N-naphthylcarbamoyl)(4,5,6,7-tetrahydrobenzo [β]thiophen-2-yl)]acetamide}. (D) Virus yield-based dose-response assays. Virus titers were determined through plaque assay (IAV-WSN, MuV F, and RSV-A2), TaqMan genome copy number quantification (IAV-Texas and IAV-Aichi), or TCID50 titration (recMeV) and then normalized to vehicle (DMSO) controls. Data points represent means for three experiments. EC50s were calculated based on four-parameter variable-slope nonlinear regression models. Specificity indices (SI) represent CC50/EC50.