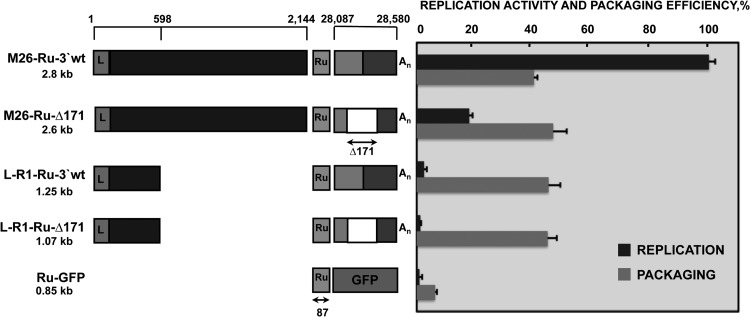
Fig 4.
Requirement of replication for packaging of M26- and R1-derived defective minigenomes. A schematic comparison of the structures of defective minigenomes analyzed in replication and packaging assays is represented on the left. Positions in the TGEV genome of the sequences included in defective minigenomes are indicated with numbers on the top. The size of each defective minigenome is indicated below its name. L, leader sequence; Ru, Rubisco sequence used for detection by qPCR. Size in nucleotides is indicated below the arrow. An, poly(A). The last 494 nt of the 3′ end of the TGEV genome are divided into two boxes, representing the 7 gene (light gray box) and the 3′ UTR (dark gray box). The white box represents the Δ171 deletion (nt 28388 to 28559) in the 3′ end of the TGEV genome, including sequences required for replication. To the right of the figure is represented the analysis by qPCR of the defective minigenome replication activity and packaging efficiencies, calculated as described in Materials and Methods. The data are the averages from five independent experiments, and error bars represent standard deviations.