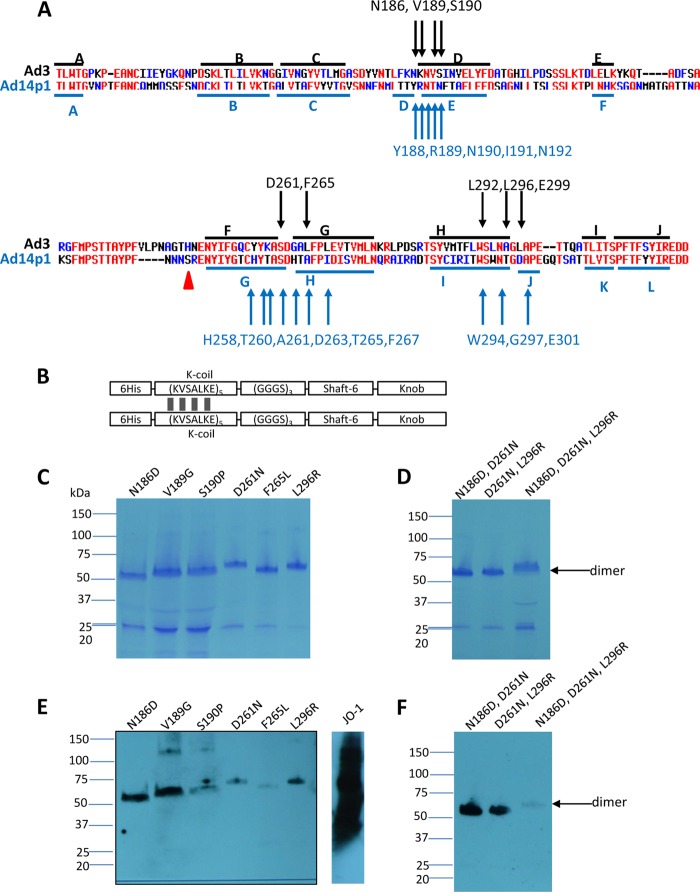
Fig 1.
Residues found to be critically involved in binding to DSG2. (A) Shown are the amino acid sequences of the Ad3 and Ad14p1 fiber knob. Beta sheets present in the Ad3 knob (PDB code 1H7Z_A) and Ad14 knob (PDB code 3F0Y_A) are indicated by blue lines. Black arrows indicate residues within the Ad3 fiber knob which, when mutated individually, ablate or reduce binding to DSG2. Compared to the parental strain of Ad14 (deWit), Ad14p1 had a deletion of two amino acid residues within the FG loop of the fiber protein knob (45), indicated by a red triangle. (B) Schematic structure of dimeric Ad3 fiber knob mutants. The fiber knob domain and one shaft motif was fused through a flexible linker to a homodimerizing K-coil domain (2). The proteins are self-dimerizing and can be purified by His-Ni-NTA affinity chromatography. (C to F) Analysis of binding of dimeric Ad3 fiber knob mutants to soluble DSG2. (C and D) Coomassie staining. Ten μg of purified Ad3 fiber knob (unboiled) was loaded per lane. Trimeric forms of the fiber knobs are indicated by an arrow. The gel contained SDS and the loading buffer containing DTT, which caused the disassembly of dimers of trimeric fiber knobs, as previously reported (2). (E and F) Western blot using soluble recombinant DSG2 as a probe, followed by anti-DSG2-MAb and anti-mouse IgG-HRP. For comparison, JO-1 (0.5 μg/lane) is shown. The Western blots were scanned and signals were quantified.