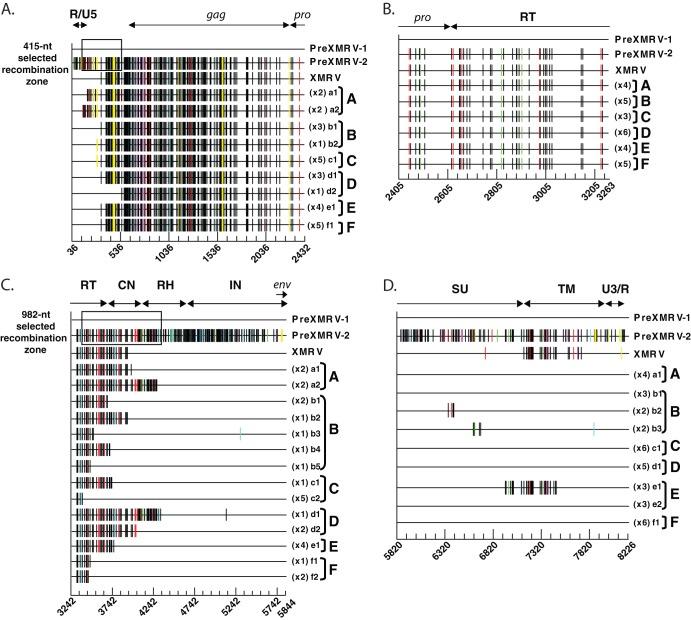
Fig 5.
Sequencing of recombinant clones. Proviral clones from infections A to F were sequenced in four regions: R/U5 to protease (A), protease to the 5′ half of pol (B), the 3′ half of pol to the end of IN (C), and the end of IN to U3/R (D). For ease of comparison, each sequence is represented as Hypermut plots; colored vertical hash marks represent nucleotide differences from PreXMRV-1, as defined by Hypermut (red, GG → GA; cyan, GA → AA; green, GC → AC; magenta, GT → AT; black, no G → A transition; yellow, gaps). The PreXMRV-1 sequence is depicted as a solid black line. For each infection, the different proviral clones (lowercase letters) and the number of each type of clone are shown. The nucleotide sequence is numbered according to the PreXMRV-1 sequence (GenBank accession number NC_007815) when aligned against PreXMRV-2 (GenBank accession number FR871850). The 415-nt and 982-nt selected recombination zone used by the RCRs in the UTR (A) and pol domain (B), respectively, are shown as a black box. The relative positions of R/U5, gag, protease (pro), reverse transcriptase (RT), connection subdomain (CN), RNase H (RH), integrase (IN), envelope (env), envelope surface unit (SU) glycoprotein, envelope transmembrane subunit (TM), and U3/R are labeled. Sequences of clones presented in this figure were deposited in GenBank.