Abstract
Here, we show that human parainfluenza viruses and Sendai virus (SeV), like other respiratory viruses, use TMPRSS2 for their activation. The membrane fusion proteins of respiratory viruses often possess serine and glutamine residues at the P2 and P3 positions, respectively, but these residues were not critical for cleavage by TMPRSS2. However, mutations of these residues affected SeV growth in specific epithelial cell lines, suggesting the importance of these residues for SeV replication in epithelia.
TEXT
In general, enveloped viruses initiate their infection by attaching to receptors on host cells, with subsequent membrane fusion between the virus envelope and the host cell membrane. The fusion process is mediated by a specialized surface glycoprotein, which is often synthesized as an inactive precursor and undergoes endoproteolysis by a host cell protease for activation. Recent studies have suggested that some type II transmembrane serine proteases (TTSPs), including TMPRSS2, human airway trypsin-like protease (HAT), TMPRSS4, and matriptase, are responsible for cleavage of the hemagglutinin (HA) protein of influenza A virus (IAV) in the human airway (1–6). Among these, TMPRSS2 has also been shown to proteolytically activate the F protein of human metapneumovirus (HMPV) (7). Severe acute respiratory syndrome (SARS) coronavirus (CoV), NL63 CoVs, and the novel human CoV (HCoV) EMC also use TMPRSS2 for their spike protein activation (8–13). Here, we studied the activation of respiratory parainfluenza viruses (human parainfluenza viruses [HPIVs] and Sendai virus [SeV]) by TMPRSS2.
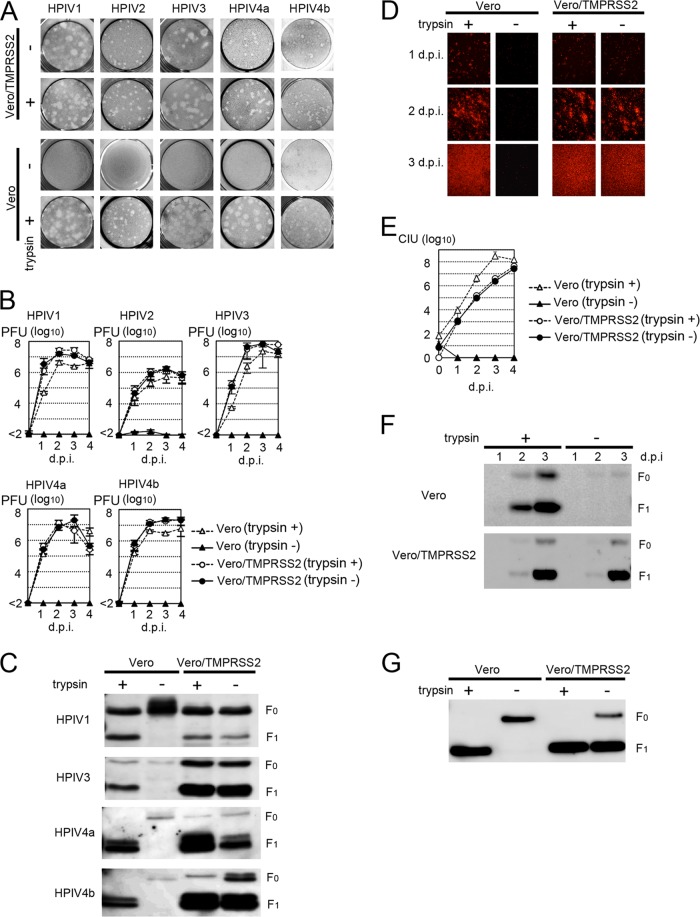
HPIV1, HPIV4a, and HPIV4b generally require trypsin (trypsin dependent) to undergo multiple rounds of infection in most established cell lines, whereas some strains of HPIV2 and HPIV3 spread efficiently in cell lines in the absence of trypsin (trypsin independent) because they use the ubiquitous furin protease (14). HPIV1 (2272-Yamagata-2009 strain), HPIV4a (M-25 strain), HPIV4b (CH19503 strain), and trypsin-dependent HPIV2 (2331-Yamagata-2009 strain) and HPIV3 (1835-Yamagata-2009 strain) were used in the present study. In the absence of trypsin, none of these HPIV strains produced plaques in Vero cells, whereas all HPIVs showed plaque formation clearly in Vero/TMPRSS2 cells, which constitutively express TMPRSS2 (Fig. 1A). In a previous study (7), we generated Vero/TMPRSS2 cells by cotransfecting Vero cells with a TMPRSS2 expression vector, pCA7-TMPRSS2, in which EcoRI and NotI sites were used for cloning, and the neo gene-bearing vector pCXN2. For multistep growth experiments, Vero/TMPRSS2 and Vero cells were infected with HPIVs at a multiplicity of infection (MOI) of 0.01. All HPIVs replicated efficiently in Vero/TMPRSS2 cells, even in the absence of trypsin (Fig. 1B). Analyses by SDS-PAGE followed by immunoblotting demonstrated cleavage of the F proteins of HPIVs in Vero/TMPRSS2 cells, but not in Vero cells, in the absence of trypsin (Fig. 1C). In these assays, rabbit antisera raised against peptides corresponding to the cytoplasmic regions of the HPIV1, HPIV3, and HPIV4 F proteins (VRRLLVMINSTNNSPINAYTLESRMRNPYM, IIIIAVKYYRIQKRNRVDQNDKPYVLTNK, and EVKNVARNQRLNRDADLFYKIPSQIPVPR, respectively) and a peroxidase-conjugated secondary antibody were used. Polyvinylidene difluoride (PVDF) membranes, on which polypeptides from cell lysates were blotted, were treated with the ECL Plus reagent (Amersham Biosciences), and chemiluminescence signals on the membrane were detected and visualized with a VersaDoc 3000 imager (Bio-Rad, Hercules, CA) (Fig. 1C). For analysis of SeV, a recombinant wild-type SeV expressing red fluorescent protein [SeV-RFP(wt)] was used. To generate SeV-RFP(wt), the RFP gene was inserted into the 3′-end-proximal first locus of the SeV genome, which is encoded by an expression plasmid, generating the full-length SeV genome plasmid pSeV-RFP (15, 16). SeV-RFP(wt) was then generated by using a reverse-genetics technique (15). In the absence of trypsin, SeV-RFP(wt) failed to undergo multiple rounds of infection in Vero cells, whereas it propagated efficiently in Vero/TMPRSS2 cells (Fig. 1D and E). Analyses by SDS-PAGE and immunoblotting also demonstrated cleavage of the F protein of SeV-RFP(wt) in Vero/TMPRSS2 cells, but not in Vero cells, in the absence of trypsin (Fig. 1F). In this assay, a rabbit antiserum raised against the cytoplasmic region of the SeV F protein (RIPRDTYTLEPKIRHMYTNGGFDAMAEKR) was used. Consistently with these data, SeV particles released from Vero/TMPRSS2 cells possessed cleaved F proteins, even in the absence of trypsin (Fig. 1G). On the other hand, SeV particles released from Vero cells cultured in the absence of trypsin possessed uncleaved F proteins (Fig. 1G). These virus particles with uncleaved F proteins were incapable of entering cells, even when the target cells expressed TMPRSS2 (data not shown). Thus, TMPRSS2 did not activate the F protein of the incoming SeV particles, as was observed for IAV (17). These data demonstrated that TMPRSS2 proteolytically activated the SeV F protein intracellularly during the process of SeV assembly. Therefore, the mechanism for SeV and IAV activation by TMPRSS2 is different from that for the SARS coronavirus, which is activated at the entry step after receptor binding (11).
Fig 1.
Proteolytic activation of HPIVs and SeV by TMPRSS2. (A) Plaque formation of HPIVs in Vero and Vero/TMPRSS2 cells in the presence or absence of trypsin. (B) Replication kinetics of HPIVs in Vero and Vero/TMPRSS2 cells. Cells were infected with HPIVs at an MOI of 0.01, cultured in the presence or absence of trypsin, and examined for their virus titers (PFU) daily. d.p.i., days postinfection. (C) Detection of HPIV F proteins by immunoblotting. Vero and Vero/TMPRSS2 cells infected with HPIVs in the presence or absence of trypsin were lysed in radioimmunoprecipitation assay (RIPA) buffer and subjected to SDS-PAGE and immunoblotting for detection of the F proteins (indicated as F0 and F1). (D) Detection of SeV-RFP(wt)-infected cells. Monolayers of Vero and Vero/TMPRSS2 cells were infected with SeV-RFP(wt) at an MOI of 0.01 and observed daily using a fluorescence microscope. (E) Replication kinetics of SeV-RFP(wt) in Vero and Vero/TMPRSS2 cells. Cells were infected with SeV-RFP(wt) at an MOI of 0.01, cultured in the presence or absence of trypsin, and examined for their virus titers (cell infectious units [CIU]) daily. (F) Detection of SeV F proteins in infected cells by immunoblotting. Vero and Vero/TMPRSS2 cells infected with SeV-RFP(wt) in the presence or absence of trypsin were lysed in RIPA buffer and subjected to SDS-PAGE and immunoblotting for detection of the F proteins. (G) Detection of SeV F proteins in virus particles by immunoblotting. Vero and Vero/TMPRSS2 cells were infected with SeV-RFP(wt) in the presence or absence of trypsin, and virus particles released from these cells were purified and subjected to SDS-PAGE and immunoblotting for detection of the F proteins.
The F proteins of HMPV, SeV, HPIV1, and HPIV4 (both HPIV4a and HPIV4b) and the HA protein of IAV (H1 subtype) possess serine and glutamine residues at the P2 and P3 positions, respectively (Table 1). These residues are relatively conserved among respiratory viruses (Table 1). An arginine or a lysine residue at P1 is mandatory for serine protease substrates, while the residues at P2 and P3 are much more flexible than the P1 residue, although they may also modulate the specificity of the protease substrate (18, 19). By site-directed mutagenesis of pSeV-RFP, five predicted amino acid substitutions (Q114A, Q114S, Q114V, S115R, and S115V) were individually introduced into the F protein of recombinant SeV. The mutated hexanucleotide sequences, which correspond to amino acid residues at positions 114 and 115 of the Q114A, Q114S, Q114V, S115R, and S115V mutants, were GCCTCG, AGCTCG, GTGTCG, ATCCGC, and ATCGTG, respectively (mutated nucleotides are underlined). Infectious SeV mutants were generated by using a reverse-genetics technique (15). The replication capacities of the mutant SeVs were analyzed using TMPRSS2-expressing Huh7 cells (Huh7/TMPRSS2-18 and Huh7/TMPRSS2m-4 cell lines) (M. Esumi et al., submitted for publication). Huh7/TMPRSS2-18 cells constitutively express an intact and active form of TMPRSS2, while Huh7/TMPRSS2m-4 cells constitutively express an enzymatically inactive mutant form of TMPRSS2 (TMPRSS2m) with an S441A mutation (Esumi et al., submitted). In Huh7/TMPRSS2m-4 cells, all of the SeVs, including SeV-RFP(wt), spread slowly, whereas in Huh7/TMPRSS2-18 cells, all of the viruses spread much more efficiently (Fig. 2A and B). However, in Huh7/TMPRSS2-18 cells, the growth of the mutant SeV with S115R (SeV-RFP/S115R) was severely deteriorated, and the infectious-virus production was more than 1,000 times lower than that of SeV-RFP(wt) (Fig. 2A and B). The other four mutants (SeV-RFP/Q114S, -RFP/Q114A, -RFP/Q114V, and -RFP/S115V) also showed impaired replication capacities that were reduced by ∼10-fold (Fig. 2B). However, pulse-chase labeling followed by immunoprecipitation and SDS-PAGE showed little, if any, effects of the mutations on the cleavage of the F proteins of the recombinant SeVs in Huh7/TMPRSS2-18 cells (Fig. 2C).
Table 1.
Residues of the cleavage site in virus membrane fusion proteins
| Virus | Residue(s) ata: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P7 | P6 | P5 | P4 | P3 | P2 | P1 | P′1 | P′2 | P′3 | |
| HMPV | E | N | P | R | Q | S | R | F | V | L |
| SeV | A | D/G | A/V | P | Q | S | R | F | F | G |
| HPIV1 | A/N | D | N/V | P | Q | S/T | R | F | F | G |
| HPIV2 (trypsin dependent) | T | A/K/T | P/T | R | Q | E | R | F | A | G |
| HPIV2 (trypsin independent) | T | K/T | T | R | Q | K | R | F | A | G |
| HPIV3 (trypsin dependent) | T | D | P | R | T | E | R | F | F | G |
| HPIV3 (trypsin independent) | T | D/N | P | R/T | T | K/R | R | F | F | G |
| HPIV4a | S | S | E | I | Q | S | R | F | F | G |
| HPIV4b | S | S | E | I | Q | S | R | F | F | G |
| IAV H1 subtype | I/V | P | S | I | Q | S | R | G | L | F |
| IAV H2 subtype | I/V | P | Q | I | E | S | R | G | L | F |
| IAV H3 subtype | I/V | P | E | K | Q | T | R | G | I/L | F |
| IAV H4 subtype | I | P | E | K | A | T | R | G | L | F |
| IAV H5 subtype | V | P | Q | R | E/K | T | R | G | L | F |
| IAV H6 subtype | I/V | P | Q | I | E | T | R | G | L | F |
| IAV H7 subtype | P | E | I | P | K | G | R | G | L | F |
| IAV H8 subtype | T | P | S | I/V | E | P | K/R | G | L | F |
| IAV H9 subtype | V | P | A | A/V | S | D | R | G | L | F |
| IAV H10 subtype | P | E | I | M | Q | G | R | G | L | F |
| IAV H11 subtype | V | P | A | I | A | S/T | R | G | L | F |
| IAV H12 subtype | V | P | Q | A/V | Q | D/N | R | G | L | F |
| IAV H13 subtype | V | P | A/T | I | A/S | N/S | R | G | L | F |
| IAV H14 subtype | I | P | G | K | Q | A | K | G | L | F |
| IAV H15 subtype | P | E | K | I | R | T | R | G | L | F |
| IAV H16 subtype | V | P | S | I/V | G/N/S/V | E | R | G | L | F |
| NDV (avirulent) | G | E/G | G | K/R | Q | G | R | L | I | G |
| Avian paramyxovirus type 2 | F | D | K | P | A | S | R | F | V | G |
| Avian paramyxovirus type 3 | Q | A/P | R | P | R/S | G | R | L | F | G |
| Avian paramyxovirus type 4 | D/E | A/V | D | I | Q | P | R | F | I | G |
| Avian paramyxovirus type 6 | H/N | P/S | A/I | P/R | E | P | R | L | I/V | G |
Serine and glutamine residues at P2 and P3 are underlined.
Fig 2.
Analyses of recombinant SeV-RFP strains with mutations at P2 and P3 of the F protein. (A) Detection of wt- and mutant-SeV-RFP-infected cells. Monolayers of Huh7/TMPRSS2-18 and Huh7/TMPRSS2m-4 cells were infected with wt and mutant SeV-RFPs at an MOI of 0.01, cultured in the absence of trypsin, and observed daily using a fluorescence microscope. Data at 3, 5, 7, and 9 days postinfection are shown. (B) Replication kinetics of wt and mutant SeV-RFPs in Huh7/TMPRSS2-18 and Huh7/TMPRSS2m-4 cells. Cells were infected with wt and mutant SeV-RFPs at an MOI of 0.01, cultured in the absence of trypsin, and examined for their virus titers (CIU) daily. (C) Pulse-chase labeling, immunoprecipitation, and SDS-PAGE for detection of SeV F proteins. Huh7/TMPRSS2-18 and Huh7/TMPRSS2m-4 cells infected with wt or mutant SeV-RFP were pulse-labeled with [35S]methionine for 15 min, cultured (chased) in normal medium for 30 or 120 min, and subjected to immunoprecipitation and SDS-PAGE for detection of the F proteins. (D) Detection of wt- and mutant-SeV-RFP-infected cells. Monolayers of Calu-3 and Caco-2 cells were infected with wt and mutant SeV-RFPs at an MOI of 0.001, cultured in the absence of trypsin, and observed daily using a fluorescence microscope. Data at 3, 5, and 6 days postinfection are shown. (E) Virus production in Calu-3 cells. Calu-3 cells were infected with wt and mutant SeV-RFPs at an MOI of 0.001 and examined for their virus titers (CIU) at 5 days postinfection.
Human bronchial epithelial Calu-3 cells and intestinal epithelial Caco-2 cells have an intrinsic ability to proteolytically activate IAV using endogenously expressed proteases (20–22). In Calu-3 cells, TMPRSS2 and matriptase were shown to contribute to IAV HA cleavage (6, 21). In Caco-2 cells, TMPRSS2 and TMPRSS4 are responsible for activation of the IAV HA (22). Calu-3 and Caco-2 cells were infected with SeV-RFP(wt) and mutant SeVs. Both cell lines supported SeV-RFP(wt) multiplication well (Fig. 2D), similar to what occurred with IAV (20–22). All of the mutant SeVs also underwent multiple rounds of infection in both cell lines. However, unlike with Huh7/TMPRSS2-18 cells, the propagation of SeV-RFP/Q114V was severely deteriorated in these cell lines, whereas SeV-RFP/S115R spread well in these cell lines (Fig. 2D and E). Although not as dramatically, the other two mutants with mutations at P3 (SeV-RFP/Q114S and -RFP/Q114A) also spread less efficiently than SeV-RFP(wt) in these cell lines (Fig. 2D and E). Thus, the effects of each mutation observed in Calu-3 and Caco-2 cells were different from those observed in TMPRSS2-expressing Huh7 cells. Since Calu-3 and Caco-2 cells have more than one protease contributing to IAV HA activation, the SeV F protein may be activated mainly by proteases other than TMPRSS2 in these cells. Nevertheless, the glutamine residue at P3 was shown to be important for SeV multiplication in bronchial and intestinal epithelial cells.
The present study has demonstrated the importance of the P2 serine and P3 glutamine residues for SeV replication, but the roles of these residues remain unclear. Although our data showed little, if any, effects of the mutations on F protein cleavage by TMPRSS2, these residues may modulate the protease's specificity or sensitivity for the F protein. Many proteases have been shown to activate respiratory viruses. Trypsin is commonly used for the propagation of IAV, HMPV, SeV, HPIVs, and coronaviruses in cultured cells. Mini-plasmin found in bronchial epithelia (23), tryptase Clara from rat lungs (24, 25), mast cell tryptase from porcine lungs (26), and factor Xa from chicken allantoic fluid (26) were shown to proteolytically activate IAV and SeV. Plasmin also activates IAV through unique mechanisms by which plasminogen is captured by the neuraminidase surface glycoprotein or host cell annexin II is incorporated into virions (27–29). Recent studies have added a new class of proteases, transmembrane serine proteases (TTSPs), to the list of IAV- or SeV-activating proteases (2). TMPRSS2, HAT (TMPRSS11D), and TMPRSS4 were shown to activate seasonal IAV strains (3, 4, 21, 22). However, it remains totally unknown which proteases contribute mainly to respiratory virus pathogenesis. Although multiple proteases may contribute to the virus' spread in vivo, membrane-anchored proteases have an advantage for processing target substrates at a specific location with a minimum concentration (30, 31). Indeed, our previous study demonstrated that a marginal level of TMPRSS2 expression was sufficient for intracellular cleavage of the IAV HA and HMPV F proteins (7). To date, the biological and physiological functions of TMPRSS2 have been poorly elucidated. Although a previous study suggested that TMPRSS2 regulates the Na+ current (32), the physiological importance of this is uncertain, because TMPRSS2 knockout mice showed no developmental, physiological, or pathological changes from wild-type mice (33). A final conclusion awaits in vivo analyses for the contribution of TMPRSS2 to viral pathogenesis. However, the accumulated evidence has revealed that TMPRSS2 acts as an activating protease for a broad range of respiratory viruses.
ACKNOWLEDGMENTS
We thank N. Ito and M. Sugiyama for providing the BHK/T7-9 cells. We also thank all the members of Department of Virology 3, NIID, Japan, for technical support.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology and the Ministry of Health, Labor and Welfare of Japan and by a grant from The Takeda Science Foundation.
Footnotes
Published ahead of print 21 August 2013
REFERENCES
- 1.Kido H, Okumura Y, Takahashi E, Pan HY, Wang S, Chida J, Le TQ, Yano M. 2008. Host envelope glycoprotein processing proteases are indispensable for entry into human cells by seasonal and highly pathogenic avian influenza viruses. J. Mol. Genet. Med. 3:167–175 [PMC free article] [PubMed] [Google Scholar]
- 2.Choi SY, Bertram S, Glowacka I, Park YW, Pohlmann S. 2009. Type II transmembrane serine proteases in cancer and viral infections. Trends Mol. Med. 15:303–312 [DOI] [PubMed] [Google Scholar]
- 3.Chaipan C, Kobasa D, Bertram S, Glowacka I, Steffen I, Tsegaye TS, Takeda M, Bugge TH, Kim S, Park Y, Marzi A, Pohlmann S. 2009. Proteolytic activation of the 1918 influenza virus hemagglutinin. J. Virol. 83:3200–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bottcher E, Matrosovich T, Beyerle M, Klenk HD, Garten W, Matrosovich M. 2006. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 80:9896–9898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamilton BS, Gludish DW, Whittaker GR. 2012. Cleavage activation of the human-adapted influenza virus subtypes by matriptase reveals both subtype and strain specificities. J. Virol. 86:10579–10586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beaulieu A, Gravel E, Cloutier A, Marois I, Colombo E, Desilets A, Verreault C, Leduc R, Marsault E, Richter MV. 2013. Matriptase proteolytically activates influenza virus and promotes multicycle replication in the human airway epithelium. J. Virol. 87:4237–4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shirogane Y, Takeda M, Iwasaki M, Ishiguro N, Takeuchi H, Nakatsu Y, Tahara M, Kikuta H, Yanagi Y. 2008. Efficient multiplication of human metapneumovirus in Vero cells expressing the transmembrane serine protease TMPRSS2. J. Virol. 82:8942–8946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertram S, Glowacka I, Muller MA, Lavender H, Gnirss K, Nehlmeier I, Niemeyer D, He Y, Simmons G, Drosten C, Soilleux EJ, Jahn O, Steffen I, Pohlmann S. 2011. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J. Virol. 85:13363–13372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glowacka I, Bertram S, Muller MA, Allen P, Soilleux E, Pfefferle S, Steffen I, Tsegaye TS, He Y, Gnirss K, Niemeyer D, Schneider H, Drosten C, Pohlmann S. 2011. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 85:4122–4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S, Gallagher T. 2011. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 85:873–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, Taguchi F. 2010. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 84:12658–12664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawase M, Shirato K, van der Hoek L, Taguchi F, Matsuyama S. 2012. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 86:6537–6545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gierer S, Bertram S, Kaup F, Wrensch F, Heurich A, Kramer-Kuhl A, Welsch K, Winkler M, Meyer B, Drosten C, Dittmer U, von Hahn T, Simmons G, Hofmann H, Pohlmann S. 2013. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2 and is targeted by neutralizing antibodies. J. Virol. 87:5502–5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karron RA, Collins PL. 2007. Parainfluenza viruses, p 1497–1526 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 15.Kato A, Sakai Y, Shioda T, Kondo T, Nakanishi M, Nagai Y. 1996. Initiation of Sendai virus multiplication from transfected cDNA or RNA with negative or positive sense. Genes Cells 1:569–579 [DOI] [PubMed] [Google Scholar]
- 16.Hasan MK, Kato A, Shioda T, Sakai Y, Yu D, Nagai Y. 1997. Creation of an infectious recombinant Sendai virus expressing the firefly luciferase gene from the 3′ proximal first locus. J. Gen. Virol. 78(Part 11):2813–2820 [DOI] [PubMed] [Google Scholar]
- 17.Bottcher-Friebertshauser E, Freuer C, Sielaff F, Schmidt S, Eickmann M, Uhlendorff J, Steinmetzer T, Klenk HD, Garten W. 2010. Cleavage of influenza virus hemagglutinin by airway proteases TMPRSS2 and HAT differs in subcellular localization and susceptibility to protease inhibitors. J. Virol. 84:5605–5614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gosalia DN, Salisbury CM, Ellman JA, Diamond SL. 2005. High throughput substrate specificity profiling of serine and cysteine proteases using solution-phase fluorogenic peptide microarrays. Mol. Cell. Proteomics 4:626–636 [DOI] [PubMed] [Google Scholar]
- 19.Gosalia DN, Salisbury CM, Maly DJ, Ellman JA, Diamond SL. 2005. Profiling serine protease substrate specificity with solution phase fluorogenic peptide microarrays. Proteomics 5:1292–1298 [DOI] [PubMed] [Google Scholar]
- 20.Zhirnov O, Klenk HD. 2003. Human influenza A viruses are proteolytically activated and do not induce apoptosis in CACO-2 cells. Virology 313:198–212 [DOI] [PubMed] [Google Scholar]
- 21.Bottcher-Friebertshauser E, Stein DA, Klenk HD, Garten W. 2011. Inhibition of influenza virus infection in human airway cell cultures by an antisense peptide-conjugated morpholino oligomer targeting the hemagglutinin-activating protease TMPRSS2. J. Virol. 85:1554–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertram S, Glowacka I, Blazejewska P, Soilleux E, Allen P, Danisch S, Steffen I, Choi SY, Park Y, Schneider H, Schughart K, Pohlmann S. 2010. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J. Virol. 84:10016–10025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murakami M, Towatari T, Ohuchi M, Shiota M, Akao M, Okumura Y, Parry MA, Kido H. 2001. Mini-plasmin found in the epithelial cells of bronchioles triggers infection by broad-spectrum influenza A viruses and Sendai virus. Eur. J. Biochem. 268:2847–2855 [DOI] [PubMed] [Google Scholar]
- 24.Kido H, Yokogoshi Y, Sakai K, Tashiro M, Kishino Y, Fukutomi A, Katunuma N. 1992. Isolation and characterization of a novel trypsin-like protease found in rat bronchiolar epithelial Clara cells. A possible activator of the viral fusion glycoprotein. J. Biol. Chem. 267:13573–13579 [PubMed] [Google Scholar]
- 25.Tashiro M, Yokogoshi Y, Tobita K, Seto JT, Rott R, Kido H. 1992. Tryptase Clara, an activating protease for Sendai virus in rat lungs, is involved in pneumopathogenicity. J. Virol. 66:7211–7216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Shiota M, Ohuchi M, Towatari T, Tashiro J, Murakami M, Yano M, Yang B, Kido H. 2000. Mast cell tryptase from pig lungs triggers infection by pneumotropic Sendai and influenza A viruses. Purification and characterization. Eur. J. Biochem. 267:3189–3197 [DOI] [PubMed] [Google Scholar]
- 27.Goto H, Kawaoka Y. 1998. A novel mechanism for the acquisition of virulence by a human influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 95:10224–10228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lazarowitz SG, Goldberg AR, Choppin PW. 1973. Proteolytic cleavage by plasmin of the HA polypeptide of influenza virus: host cell activation of serum plasminogen. Virology 56:172–180 [DOI] [PubMed] [Google Scholar]
- 29.LeBouder F, Morello E, Rimmelzwaan GF, Bosse F, Pechoux C, Delmas B, Riteau B. 2008. Annexin II incorporated into influenza virus particles supports virus replication by converting plasminogen into plasmin. J. Virol. 82:6820–6828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hooper JD, Clements JA, Quigley JP, Antalis TM. 2001. Type II transmembrane serine proteases. Insights into an emerging class of cell surface proteolytic enzymes. J. Biol. Chem. 276:857–860 [DOI] [PubMed] [Google Scholar]
- 31.Antalis TM, Buzza MS, Hodge KM, Hooper JD, Netzel-Arnett S. 2010. The cutting edge: membrane-anchored serine protease activities in the pericellular microenvironment. Biochem. J. 428:325–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donaldson SH, Hirsh A, Li DC, Holloway G, Chao J, Boucher RC, Gabriel SE. 2002. Regulation of the epithelial sodium channel by serine proteases in human airways. J. Biol. Chem. 277:8338–8345 [DOI] [PubMed] [Google Scholar]
- 33.Kim TS, Heinlein C, Hackman RC, Nelson PS. 2006. Phenotypic analysis of mice lacking the Tmprss2-encoded protease. Mol. Cell. Biol. 26:965–975 [DOI] [PMC free article] [PubMed] [Google Scholar]