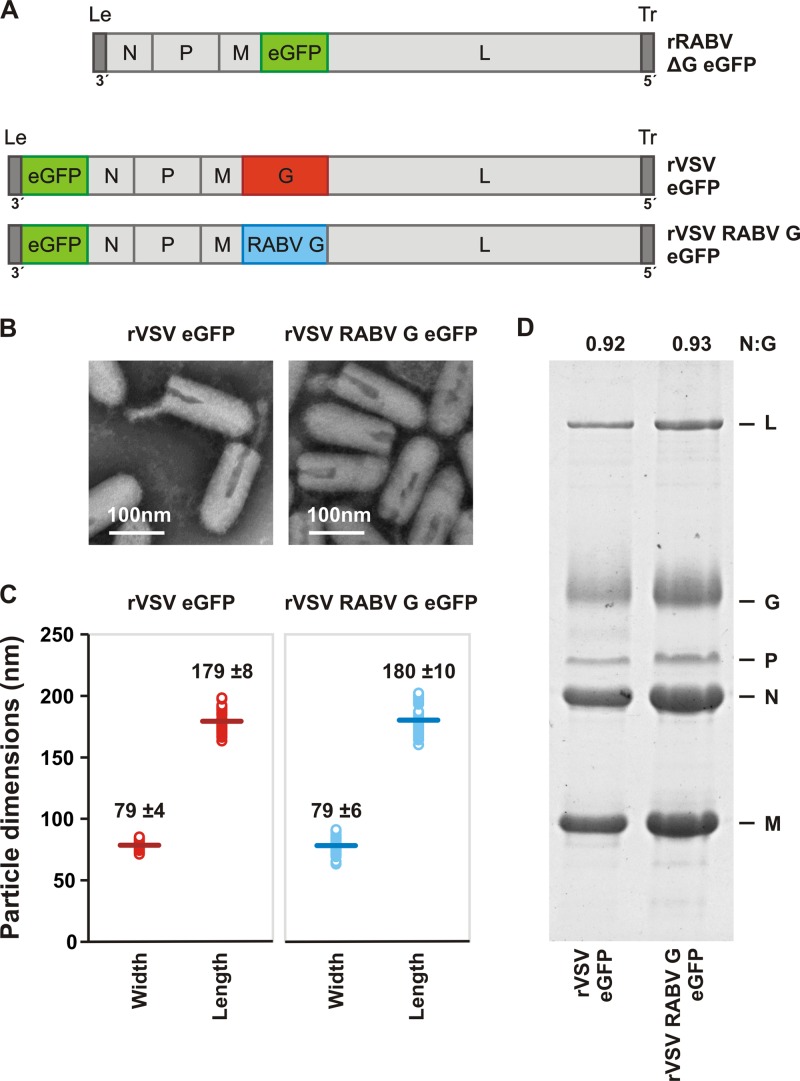
Fig 1.
Characterization of recombinant VSV expressing RABV G. (A) Genomic structures of rRABV ΔG eGFP, rVSV eGFP, and rVSV RABV G eGFP. The single-stranded, negative-sense RNA genomes are shown in a 3′ to 5′ orientation. N, nucleocapsid gene; P, phosphoprotein gene; M, matrix gene; G, glycoprotein gene; L, large polymerase gene. Noncoding genomic leader (Le) and trailer (Tr) regions serve as promoters for RNA synthesis and genomic RNA encapsidation, respectively. All viruses express the eGFP gene (green) as a marker for infection. The glycoprotein open reading frame of rRABV ΔG eGFP was replaced with that of eGFP. To produce infectious virus, rRABV ΔG was grown in complementing cells expressing SAD B19 G. In rVSV RABV G eGFP, the wild-type VSV G (1,535 nt) was replaced with RABV G (1,574 nt) from the SAD B19 strain. (B) Electron micrographs of rVSV eGFP and rVSV RABV G eGFP viral particles demonstrate morphological homogeneity. Particles were negatively stained with 1% PTA. (C) Dimensions of individual viral particles measured from micrographs like those shown in panel B. Each open circle represents the measurement for a single virion. A line denotes the mean (± the standard deviation [SD]; n = 50) for each population, the value of which is provided. (D) SDS-PAGE analysis of purified virions. Viral proteins were stained with Coomassie blue. The ratio of N to G was quantified using ImageJ to estimate the average glycoprotein density in each particle population.