Fig 1.
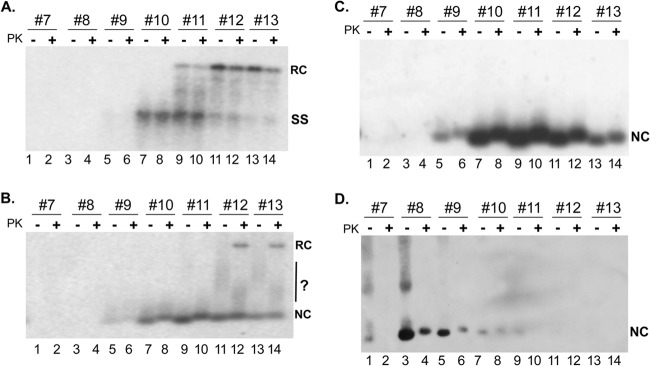
Isolation of HBV NCs from MNase-treated lysates by linear sucrose gradient centrifugation and detection of NC-associated viral DNA, RNA, and capsid protein. Induced HepAD38 cells were lysed by using NP-40, and the cytoplasmic lysate was treated with MNase. The supernatant from the MNase treatment was fractionated by linear sucrose gradient centrifugation as described in Materials and Methods. (A and B) Aliquots from fractions 7 to 13 were treated with or without PK and resolved on an agarose gel with (A) or without (B) 0.1% SDS, which was also added to the samples loaded in panel A. Viral DNA released from the SDS-disrupted NCs (A) or associated with native NCs (B) was detected by Southern blotting with an HBV-specific DNA probe. In both panels A and B, the agarose gel was treated with NaOH to remove RNA as well as to denature DS DNA. (C) To detect capsid-associated pgRNA, the NaOH treatment was omitted, and viral RNA was detected by using a riboprobe specific for the plus strand of the HBV genome. (D) Capsid protein was detected by using and anti-HBc MAb specific for the HBc N terminus (see Materials and Methods for details). NC, nucleocapsid; RC, relaxed circular DNA; SS, single-stranded DNA. The question mark in panel B denotes putative NCs with altered mobility on the gel.
