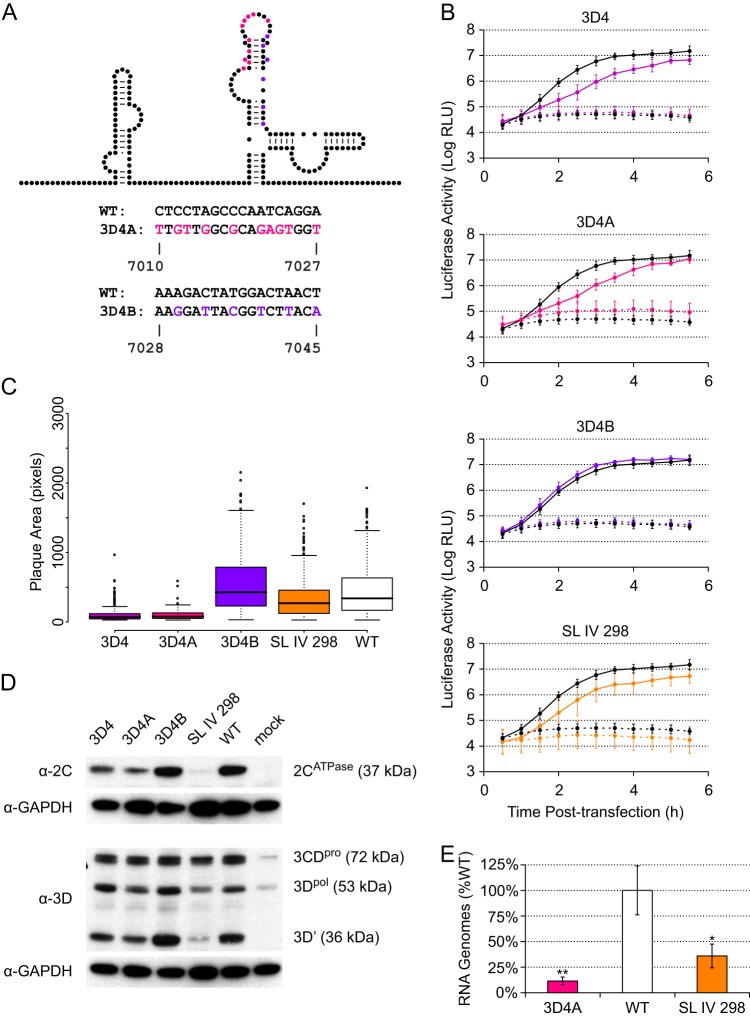
Fig 5.
Subdivision and further characterization of the 3D4 mutant. (A) Mutants. On the schematic, color indicates nucleotides targeted in each mutant. The actual nucleotide changes and corresponding wild-type sequence and nucleotide positions for each mutant are listed below. For WT structure, A-U and C-G base pairing is indicated by lines and G-U (wobble) base pairing by dots. (B) PLuc assay. Mutant (colored) or wild-type (black) replicon RNA was transfected into HeLa S3 cells at 37°C. Cells were incubated in the presence (dashed lines) or absence (solid lines) of 2 mM GdnHCl. Luciferase activity (relative light units) corresponding to ∼3.1 × 105 cells was measured every 30 min. Data are plotted as the means ± standard deviations for three independent transfections. Replication delays at 2.0 to 3.5 h posttransfection are significant compared to results for the wild type in 3D4 and 3D4A (P < 0.05). (C) Plaque assays with plaque size measurement. Clonal stocks were used to infect HeLa S3 cells at 37°C. Box plots indicate the median plaque area in pixels, 25% and 75% quartiles, and 1.5× interquartile range; outliers are shown as open circles. 3D4 (P < 2.2E−16), 3D4A (P < 2.2E−16), and SL IV 298 (P = 5.6E−05) plaques are significantly smaller than the wild type. (D) Western blots. P1 virus stocks were used to infect HeLa S3 cells (MOI of ∼20). Total protein was harvested at 5 h postinfection. Five microliters of cell lysate was run for each sample and blotted with anti-3D or anti-2C. Membranes were stripped and reblotted with anti-GAPDH. (E) qPCR. P1 virus stocks were used to infect HeLa S3 cells (MOI of ∼25). Total RNA was harvested at 5 h postinfection, and positive-sense RNA was measured by strand-specific qPCR. Data are normalized to the average wild-type value and are plotted as the means ± standard deviations for three replicates.