Abstract
Classical swine fever virus (CSFV) is a positive-stranded RNA virus belonging to the genus Pestivirus within the Flaviviridae family. Pivotal for processing of a large portion of the viral polyprotein is a serine protease activity within nonstructural protein 3 (NS3) that also harbors helicase and NTPase activities essential for RNA replication. In CSFV-infected cells, NS3 appears as two forms, a fully processed NS3 of 80 kDa and the precursor molecule NS2-3 of 120 kDa. Here we report the identification and mapping of additional autocatalytic intramolecular cleavages. One cleavable peptide bond occurs between Leu1781 and Met1782, giving rise to a helicase subunit of 55 kDa and, depending on the substrate, a NS2-3 fragment of 78 kDa (NS2-3p) or a NS3 protease subunit of 26 kDa (NS3p). In trans-cleavage assays using NS4-5 as a substrate, NS3p acts as a fully functional protease that is able to process the polyprotein. NS3p comprises the minimal essential protease, as deletion of Leu1781 results in inactivation. A second intramolecular cleavage was mapped to the Leu1748/Lys1749 peptide bond that yields a proteolytically inactive NS3 fragment. Deletion of either of the cleavage site residues resulted in a loss of RNA infectivity, indicating the functional importance of amino acid identity at the respective positions. Our data suggest that internal cleavage within the NS3 moiety is a common process that further extends the functional repertoires of the multifunctional NS2-3 or NS3 and represents another level of the complex polyprotein processing of Flaviviridae.
INTRODUCTION
Classical swine fever virus (CSFV) is the causative agent of an important disease in pigs and is listed by the World Organization for Animal Health (OIE). CSFV, together with bovine viral diarrhea virus (BVDV) and border disease virus (BDV), represents the genus Pestivirus within the Flaviviridae (1).
The genome of CSFV, a positive-stranded RNA molecule of about 12.3 kb, encodes a single polyprotein of 3,899 amino acids that is co- and posttranslationally processed into 12 mature proteins by two cellular and three viral proteases (Npro, nonstructural protein 2 [NS2], and NS3) (2, 3). Npro and NS2 are autoproteases that mediate a single cis-cleavage each. The chymotrypsin-like serine protease NS3 acts in both cis-cleavage (NS3-4A) and trans-cleavage (NS4A-4B-5A-5B) (4). In addition to its protease activity, NS3 mediates helicase (5) and NTPase (6) activities. While the C-terminal DEXH helicase (superfamily 2) shares a high degree of conservation within the members of Flaviviridae, the NS3 serine proteases are variable in sequence and substrate specificity. NS3-mediated cleavages in pestiviruses occur at Leu/Ser, Leu/Ala, and Leu/Asn sites (7), while NS3 of hepatitis C virus (HCV) prefers Cys/X motives (8) and Flavivirus NS3 dibasic motives (9). The NS3 protease of pestiviruses and hepaciviruses utilizes NS4A as an essential cofactor (4), while the NS3 protease of flaviviruses requires NS2B to form the active enzyme (9, 10). Different in vitro models have been established to study the NS3 proteases of Flaviviridae using either mammalian cell-based (4) or bacterial expression systems. The NS3 protease of HCV has been intensively studied as a target for chemotherapy of chronic HCV patients. As such, proteolytically active single-chain NS4A3 constructs of HCV have been engineered for bacterial expression, representing N-terminal fusions of a central segment of NS4A with NS3 via a short linker sequence (11, 12). For HCV, an intramolecular processing of NS3 has been reported that occurs within the helicase domain (13).
In addition to NS3, the precursor NS2-3 has been identified as a key molecule within the pestiviral life cycle regulating both particle assembly and replication (14). Uncleaved NS2-3 is essential for CSFV particle formation (15), even though a genetically engineered BVDV mutant was able to produce virus progeny in the absence of uncleaved NS2-3 precursors (16). Detailed analyses demonstrated that the availability of the cellular cofactor JIV (“J-domain protein interacting with viral protein”; DNAJC14) restricts NS2-3 cleavage to the first hours of infection in the noncytopathogenic (ncp) biotype whereas a continuous NS2-3 cleavage takes place in the cytopathogenic (cp) biotype of BVDV (17). Elevated levels of mature NS3 led to an enhanced processing of NS4-5 precursor molecules (18) and have been linked to the accelerated RNA replication of cp BVDVs (17). In a recent study, the decisive function of the NS3 helicase for virus particle formation became evident. A deletion of the essential core protein-encoding sequence was compensated for by single mutations in domain 3 of the helicase (19). Studying the protease and helicase of CSFV NS3, we identified novel specific fragments of NS2-3 and NS3 in infected cells. Intramolecular cleavage also occurred in a bacterial expression construct using a single-chain NS4A3 protease. Here we report on a novel biologically active fragment of NS3 that turns another page in the catalogue of the multiple functions of this molecule.
MATERIALS AND METHODS
Cells and viruses.
SK-6 cells (20) and BHK-21 cells (21) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS). All cells were maintained at 37°C and 5% CO2. ncp CSFV, cp CSFV-JIV, and a cp CSFV replicon were generated by transfection of a SP6 transcript of p447, p447-JIV (22), and p447-rep cDNA (18) clones as described previously (23). Nucleotide and amino acid numbers of CSFV throughout this study refer to the sequence of the parental ncp CSFV strain Alfort-Tuebingen (GenBank J04358.2). A modified vaccinia virus Ankara strain, MVA/T7 pol, was used for the mammalian expression of pCite-derived plasmids, as described previously (24).
Generation of DNA plasmids.
The plasmids used in this study were generated using standard methods as briefly described below. Primer sequences are available upon request. All mutagenized DNA plasmids were verified by nucleotide sequencing.
(i) Generation of an expression plasmid encoding a C-terminal fragment of NS3.
The coding sequences of a C-terminal fragment of NS3 were ligated into a modified pet11a vector (Merck, Darmstadt, Germany) for expression in Escherichia coli with an N-terminal hepathistidine tag (MHHHHHHH). The resulting plasmid was termed pet11a-NS3-C-term. Mutagenesis was performed by PCR with Pfu DNA polymerase (Promega, Madison, WI).
(ii) Generation of a bacterial expression plasmid encoding CSFV single-chain NS4A3.
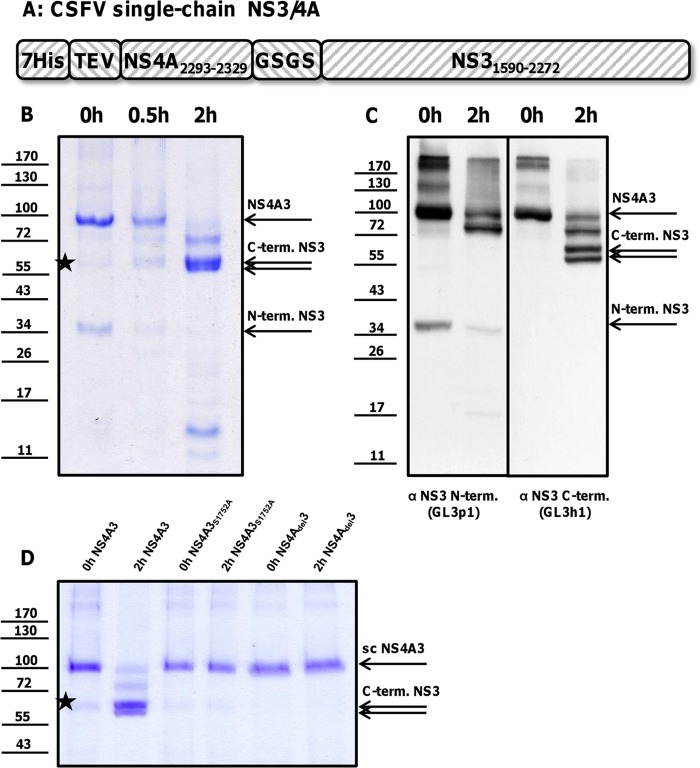
The coding sequence of the active cofactor peptide of CSFV NS4A (amino acids [aa] 2293 to 2329) was amplified by PCR and ligated into a modified pet11a vector with an N-terminal polyhistidine tag followed by a Tobacco Etch Virus (TEV) protease cleavage site (MHHHHHHHENLYFQG). A nucleotide sequence encoding a linker and the entire CSFV NS3 (aa 1590 to 2272) was amplified by PCR and inserted at the 3′ end of the active cofactor peptide of NS4A, resulting in the plasmid pet11a-scNS4A3 with a proteolytically active single-chain NS4A3 protease. The NS3 protease was inactivated by the point mutation Ser1752Ala introduced by quick-change PCR to generate the plasmid pet11a-scNS4A3S1752A. A deletion of the nucleotide sequence encoding the first amino acids of the NS4A cofactor peptide (SKRHIPVVT) in the plasmid pet11a-scNS4Adel3 was introduced to abolish the catalytic activity of the protease. A scheme of the NS4A3 expression construct is given (see Fig. 3A).
Fig 3.
(A) Internal cleavage of a CSFV NS4A3 construct. (B) NS4A3 immediately after purification by IMAC (0 h NS4A3) or after incubation for 0.5 h or 2 h at 37°C (0.5 h or 2 h NS4A3) was solved by SDS-PAGE and visualized by Coomassie staining. (C) Degradation products were specified using antibodies against the N and C termini of NS3 (the applied antibodies are indicated below the figure panel). The dominant protein bands were attributed to NS3 (75 kDa), two C-terminal fragments (58 and 55 kDa), and one short-lived N-terminal fragment (35 kDa). To inactivate the NS4A3 protease, a part of the NS4A cofactor peptide was deleted (NS4Adel3) or the essential Ser1752 was mutated (NS4A3S1752A). Inactivation of the NS4A3 protease stabilized the construct (shown in panel D). The positions of NS4A3 and processing products are indicated at the right. A weak 58-to-60-kDa band visible in all preparations (marked with stars in panels B and D) is most likely a copurified bacterial protein that is not detected by NS3-specific antibodies (C). Molecular mass standards (kDa) are shown on the left.
(iii) Generation of CSFV NS3 cleavage site mutants.
The region encoding NS2-3 of CSFV was amplified by PCR using p447 as a template. The DNA fragment was ligated into a pCite2a vector (Novagen, Madison, WI) to generate the plasmid pCite-NS2-3. All mutations were introduced by outward PCR. DNA fragments carrying the respective mutations were transferred into homologous BglII/EcoRI sites of p447 to generate the mutated full-length CSFV clones. A SalI/EcoRI fragment of these mutated p447 genomes was transferred to the homologous sites of the cp CSFV replicon clone p447rep to introduce the mutation in the replicon context.
(iv) Generation of plasmids for the transient expression of NS3 enzymes and NS4-5 substrates in mammalian cells.
The plasmids used for transient expression in mammalian cells were based on a pCite2a backbone. This vector provides a T7 polymerase promoter upstream of the internal ribosomal entry site (IRES) of encephalomyocarditis virus. We amplified the coding sequences of CSFV nonstructural proteins by PCR and inserted them into a pCite vector, as described before. The resulting plasmids were named pCite-NS2-3-4A (aa 1134 to 2336), pCite-NS2-3 (aa 1134 to 2272), pCite-NS3-4A (aa 1590 to 2336), pCite-NS3 (aa 1590 to 2272), pCite-NS4A-4B-5A-5B (aa 2273 to 3898), and pCite-NS4B-5A-5B (aa 2337 to 3898). NS2-3 cleavage was inhibited by the introduction of the mutation C1512A (25) in the plasmid pCite-NS2-3C1512A using outward PCR. C-terminally truncated NS3 constructs pCite-NS3p1748 (aa 1590 to 1748), pCite-NS3p1779 (aa 1590 to 1779), pCite-NS3p1780 (aa 1590 to 1780), pCite-NS3p1781 (aa 1590 to 1781), and pCite-NS3p1782 (aa 1590 to 1782) were generated by PCR. The plasmid pCite-NS3pL1781A is based on pCite-NS3 (full length) and carries the indicated mutation introduced by PCR. An overview of all pCite expression constructs is given in Table 1.
Table 1.
Plasmids used in this study
| Plasmid name | Description |
|---|---|
| p447 | cDNA copy of ncp CSFV, strain Alfort-Tübingen |
| p447-JIV | cDNA copy of cp CSFV, strain Alfort-Tübingen |
| p-447rep | cDNA copy of a cp CSFV replicon, strain Alfort-Tübingen |
| pet11a-NS3-C-term | Bacterial expression construct for a polyhistidine-tagged C-terminal fragment of CSFV NS3 (aa 2108–2272) |
| pet11a-scNS4A3 | Bacterial expression construct for a polyhistidine-tagged single-chain NS4A3 protease |
| pet11a-scNS4A3S1752A | Bacterial expression construct for an inactivated polyhistidine-tagged single-chain NS4A3 protease |
| pet11a-scNS4Adel3 | Bacterial expression construct for a polyhistidine-tagged single-chain NS4A3 protease lacking parts of the NS4A cofactor |
| pCite-NS2-3 | Mammalian cell expression construct for CSFV NS2-3 |
| pCite-NS2-3ΔL1748 | Mammalian cell expression construct for CSFV NS2-3 carrying the indicated mutation |
| pCite-NS2-3ΔL1781 | |
| p447ΔL1748 | cDNA copy of ncp CSFV, strain Alfort-Tübingen, carrying the indicated mutation |
| p447ΔL1781 | |
| p447L1748A | |
| p447L1748G | |
| p447L1748N | |
| p447L1748R | |
| p447L1781A | |
| p447L1781G | |
| p447L1781N | |
| p447L1781I | |
| p447L1781V | |
| p447M1782S | |
| p447repΔL1748 | cDNA copy of a cp CSFV replicon, strain Alfort-Tübingen, carrying the indicated mutation |
| p447repΔL1781 | |
| p447repL1748A | |
| p447repL1748G | |
| p447repL1748N | |
| p447repL1781A | |
| p447repL1781G | |
| p447repL1781N | |
| pCite-NS2-3-4A | Mammalian cell expression construct for CSFV NS2-3-4A |
| pCite-NS3-4A | Mammalian cell expression construct for CSFV NS3-4A |
| pCite-NS3 | Mammalian cell expression construct for CSFV NS3 |
| pCite-NS4A-4B-5A-5B | Mammalian cell expression construct for CSFV NS4A-4B-5A-5B |
| pCite-NS4B-5A-5B | Mammalian cell expression construct for CSFV NS4B-5A-5B |
| pCite-NS2-3C1512A | Mammalian cell expression construct for CSFV NS2-3 carrying the mutation C1512A which inhibit the NS2-3 cleavage |
| pCite-NS2-3-4AC1512A | Mammalian cell expression construct for CSFV NS2-3-4A carrying the mutation C1512A |
| pCite-NS3p1748 | Mammalian cell expression construct for CSFV NS3p1748 |
| pCite-NS3p1779 | Mammalian cell expression construct for CSFV NS3p1779 |
| pCite-NS3p1780 | Mammalian cell expression construct for CSFV NS3p1780 |
| pCite-NS3p1781 | Mammalian cell expression construct for CSFV NS3p1781 |
| pCite-NS3p1782 | Mammalian cell expression construct for CSFV NS3p1782 |
| pCite-NS3pL1781A | Mammalian cell expression construct for CSFV NS3p1781 carrying the amino acid exchange L1781A |
Transient expression.
A total of 1 × 106 BHK-21 cells were seeded on a 3.5-cm-diameter tissue culture dish in 2.5 ml of DMEM with 10% FCS. The cells were infected with MVA/T7 pol at a multiplicity of infection (MOI) of 10 for 4 h at 37°C. Transient expression of recombinant proteins was achieved by transfection of 2.5 μg plasmid DNA (1.25 μg each in case of double transfection) employing Superfect (Qiagen, Hilden, Germany) according to the manufacturer's recommendation. At 12 h after transfection, the cell cultures were harvested in SDS buffer for Western blot analysis.
CSFV infection, cell lysis, and indirect immunoperoxidase staining.
Transfection of SK-6 cells with infectious CSFV RNA was achieved by electroporation as described before (18). SK-6 cells were infected with ncp CSFV or cp CSFV-JIV progeny virus at an MOI of one. Protein extracts of ncp CSFV- or cp CSFV-JIV-infected cells were prepared 48 h postinfection (p.i.) and 15 h posttransfection (p.t.) of cp CSFV replicon RNA. Briefly, 1 × 106 of ncp CSFV-infected, cp CSFV-JIV-infected, cp CSFV replicon-transfected, or uninfected SK-6 cells were washed with phosphate-buffered saline (PBS) and harvested in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris, 150 mM NaCl, 0.5% sodium desoxycholate, 1% NP-40, 0.5 mM PefaBloc SC [Roche], pH 8.0). After an incubation of 1 h at 4°C, the nuclei were removed by centrifugation (5,000 × g for 5 min) and the supernatant was subjected to SDS-PAGE analysis. For proteasome inhibitor treatment, at 18 h postinfection, 50 μM MG-132, 50 μM bortezomib (Selleck Chemicals, Houston, TX), or an equal volume of dimethyl sulfoxide (DMSO) was added to the medium for 6 h. Cells were washed with PBS and harvested in RIPA buffer at 24 h p.i.
To examine the efficiency of infection following RNA transfection, we used indirect immunoperoxidase staining. Therefore, the cells were fixed with a mixture of equal amounts of ice-cold methanol and acetone for 2 min and air dried. After rehydration with PBS, the cells were incubated with monoclonal antibody (moAb) 8.12.7 (26), kindly provided by E. J. Dubovi (Cornell University, Ithaca, NY). Indirect immunoperoxidase staining was achieved using horseradish peroxidase (HRPO)-conjugated goat anti-mouse IgG (Dianova, Germany) and AEC substrate. Infected cells were counted using a Nikon Eclipse TS100 microscope.
Preparation of recombinant proteins.
The heterologous expression of a C-terminal NS3 fragment and of NS4A3 constructs was achieved via a T7 RNA polymerase promoter containing plasmid (pET11a) in E. coli strain Rosetta 2 (Merck, Darmstadt, Germany). Transformed cells were grown in Luria-Bertani medium at 37°C, and expression was induced at an optical density (OD) of 0.8 with 1 mM isopropyl-ß-d-thiogalactopyranoside (IPTG; AppliChem, Darmstadt, Germany). Expression of the recombinant proteins was performed at 30°C with vigorous aeration for 2 h. Cells were harvested by centrifugation, resuspended in lysis buffer (300 mM NaCl, 50 mM Na2PO4, 50 mM imidazole, 2 mM ß-mercaptoethanol, 0.5% [vol/vol] Triton X-100, 1% [vol/vol] His protease inhibitor mix [Sigma-Aldrich, St. Louis, MO], pH 8.0), and lysed by three cycles of freezing and thawing. After ultrasonication (1 min with external probe), the insoluble matter was removed by ultracentrifugation at 105 × g for 1 h. The proteins were purified by ion metal affinity chromatography (IMAC) with Ni2+ Sepharose (HisTrap; GE Healthcare, Port Washington, NY) and eluted by the use of an imidazole gradient (50 to 500 mM imidazole, 300 mM NaCl, 50 mM Na2PO4, pH 8.0) as recommended by the supplier. The amounts and purity of proteins were assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and identity was confirmed by Western blot analysis with an anti-histidine-tag antibody. Quantities of 0.3 mg of the C-terminal NS3 fragment and of about 0.5 mg of each of the NS4A3 constructs were obtained from 250-ml cultures. The purified C-terminal fragment of NS3 was dialyzed against PBS and used for immunization of mice. It also served as an antigen source for the enzyme-linked immunosorbent assay (ELISA) screening of monoclonal antibodies. An aliquot of all purified NS4A3 constructs was directly harvested in SDS-PAGE buffer and heat inactivated (95°C for 5 min) to serve as a reference. Another aliquot was dialyzed against PBS and used to study the autocatalytic degradation of NS4A3 proteins.
Generation of monoclonal antibodies against the C terminus of NS3.
Hybridomas secreting the desired monoclonal antibodies against the C-terminal fragment of CSFV NS3 were generated according to a modified standard procedure as described previously (18, 27). The nomenclature of hybridomas and properties of secreted moAbs used in this study is indicated in Table 2.
Table 2.
Hybridomas secreting moAbs against the C terminus of NS3
| Hybridoma | From well: | IgG subtype | Reactivity | Properties determined bya: |
|---|---|---|---|---|
| GL3h1 | 10C11 | IgG1 | NS3 helicase | WB, IF, IP |
| GL3h2 | 13B6 | IgG1 | NS3 helicase | WB, IP |
| GL3h3 | 2F9 | IgG1 | NS3 helicase | WB, IP |
WB, Western blot; IF, immunofluorescence; IP, immunoprecipitation.
SDS-PAGE and immunoblotting.
Proteins were separated in polyacrylamide-tricine gel systems (28). After SDS-PAGE, proteins were transferred to a nitrocellulose (Pall Corporation, Port Washington, NY) or polyvinylidene fluoride (Immobilon-FL; Sigma) membrane. Membranes were blocked with 4% (wt/vol) dried skim milk–PBS–0.1% (vol/vol) Tween 20. After incubation with primary antibodies, HRPO-conjugated goat anti-mouse IgG (Dianova) and a chemiluminescence reagent (ECL; PerkinElmer, Waltham, MA) were applied for signal detection.
N-terminal protein sequencing.
N-terminal sequencing of proteins was done using Edmann degradation of protein bands excised from SDS-PAGE.
RESULTS
Detection of a novel intramolecular cleavage within the NS3 of CSFV.
Recently, we reported on the time-resolved hierarchy of CSFV nonstructural protein processing (18). As a prerequisite for studying the NS2-3 cleavage in virus-infected cells, a monoclonal antibody (moAb) was established that was directed against the protease domain of NS3 (GL3p1). In Western blot and immunoprecipitation experiments, this moAb recognized two protein species of 120 and 80 kDa representing NS2-3 and NS3. In an additional approach, complementary monoclonal antibodies specific for the C terminus of NS3 (GL3h) were established (Table 2). The organization of NS2-3 of CSFV, the functional domains of NS3, and the binding positions of our moAbs are schematically depicted (see Fig. 2A).
Fig 2.
NS3 fragments detected with moAbs specific for the N and C termini of CSFV NS3. (A) Scheme of CSFV NS2-3 organization, NS2 autoprotease cleavage site, and NS3 domain structure and binding positions of the respective antibodies. (B and C) Western blot analysis of NS3 48 h after infection of SK-6 cells with ncp CSFV or cp CSFV-JIV and 15 h after transfection with RNA of a cp CSFV replicon. Mock-infected SK-6 cells served as a control. The antibody specific for the C terminus of NS3 detected a NS3 fragment with an apparent molecular mass of 55 kDa (NS3 fragment labeled NS3h in panel B). In contrast, the antibody specific for the protease domain of NS3 identified two N-terminal fragments with apparent molecular masses of 26 and 24 kDa in the cp CSFV replicon only (NS3 fragments labeled “NS3p” in panel C). (D) Proteasome inhibitor treatment of mock-, ncp CSFV-, and cp CSFV-JIV-infected SK-6 cells. NS3 was detected with the protease domain-specific antibody in the absence or presence of MG-132 (50 μM for 6 h) or bortezomib (50 μM for 6 h). A novel polyprotein fragment of 78 kDa became visible after proteasome inhibitor treatment (fragment labeled “NS2-3p” in panel D). The positions of NS2-3, NS2-3p, NS3, NS3h, and NS3p are indicated on the right. Molecular mass standards (kDa) are shown on the left.
Continuing the previous experimental approaches, the three different prototypes of pestivirus replication levels (Fig. 1), ncp CSFV (field virus), cp CSFV (JIV-enhanced field virus), and CSFV replicon (subgenomic RNA), were compared in Western blot analysis using moAb GL3h1 (Fig. 2B). In addition to NS2-3 and NS3 bands, this moAb detected a broad protein band of about 55 kDa in cells replicating ncp CSFV, cp CSFV-JIV, and the cp CSFV replicon (Fig. 2B, NS3 fragment labeled “NS3h”). In cp CSFV replicon-transfected cells, an additional weak band of 58 kDa appeared. This band is also visible in ncp and cp CSFV-infected cells upon very long exposure of the films (data not shown). Interestingly, the 55-kDa NS3 fragment has been observed before, but the exact nature of this protein remained obscure (see, e.g., Fig. 5 in reference 17). If the moAb directed against the C terminus detects a NS3 fragment that is not reactive with the moAb developed against the N terminus, there is a missing mass of about 25 kDa. This mass should locate toward the N terminus of NS3, covering most of the protease domain. Close inspection of lysates from CSFV replicon-infected cells with moAb GL3p1 revealed a doublet of 24- and 26-kDa proteins (Fig. 2C, NS3 fragment labeled “NS3p”) that was absent from CSFV- and CSFV-JIV-infected cells.
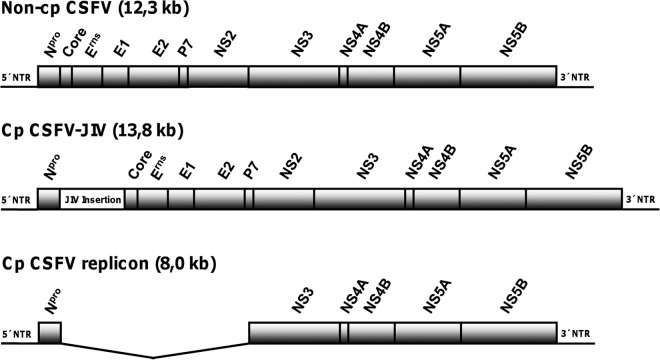
Fig 1.
Organizations of viral genomes used in this study. The three prototypical forms of pestiviral replication were represented by a wild-type non-cp CSFV clone (p447, top), its cp derivative (CSFV-JIV, center), and a cp subgenome (CSFV replicon, bottom). Relative positions of viral genes encoding the structural proteins (Core, Erns, E1, and E2) and nonstructural proteins (Npro, P7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) are indicated. Cp CSFV-JIV contains an insertion of 1.539 kb derived from cp BVDV strain CP8 (GenBank AY182137.1). The inserted sequence encodes a complex fusion protein (513 amino acids) that is composed of viral sequences (Core, Erns, and Npro) as well as cellular sequences (JIV-1, JIV-2, and bovine homologue to human nuclear protein 1 [Hcc-1]). The cp CSFV replicon lacks the coding region for Core to NS2. NTR, nontranslated region.
Fig 5.
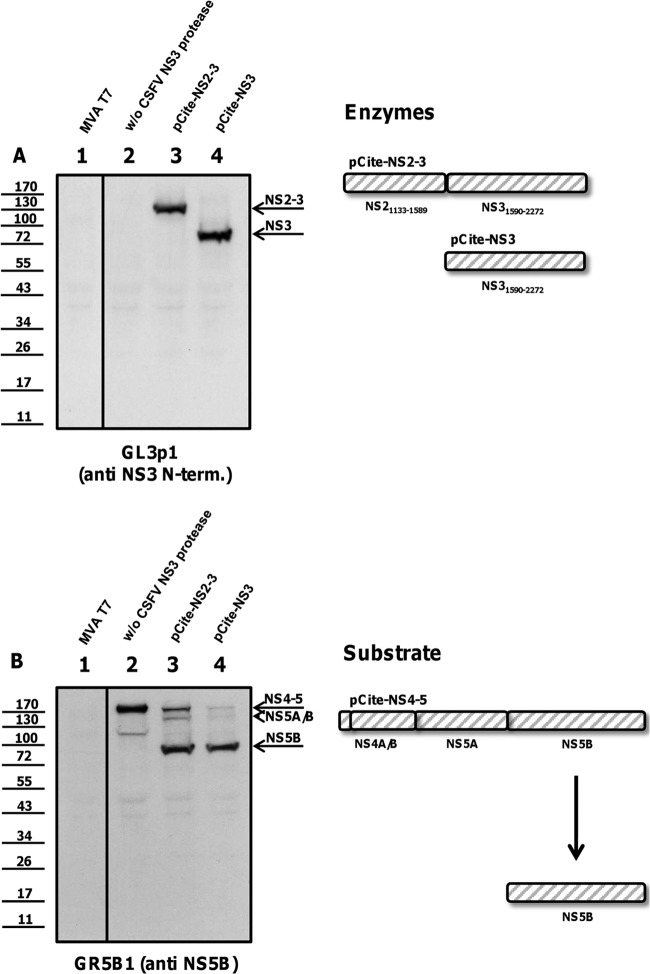
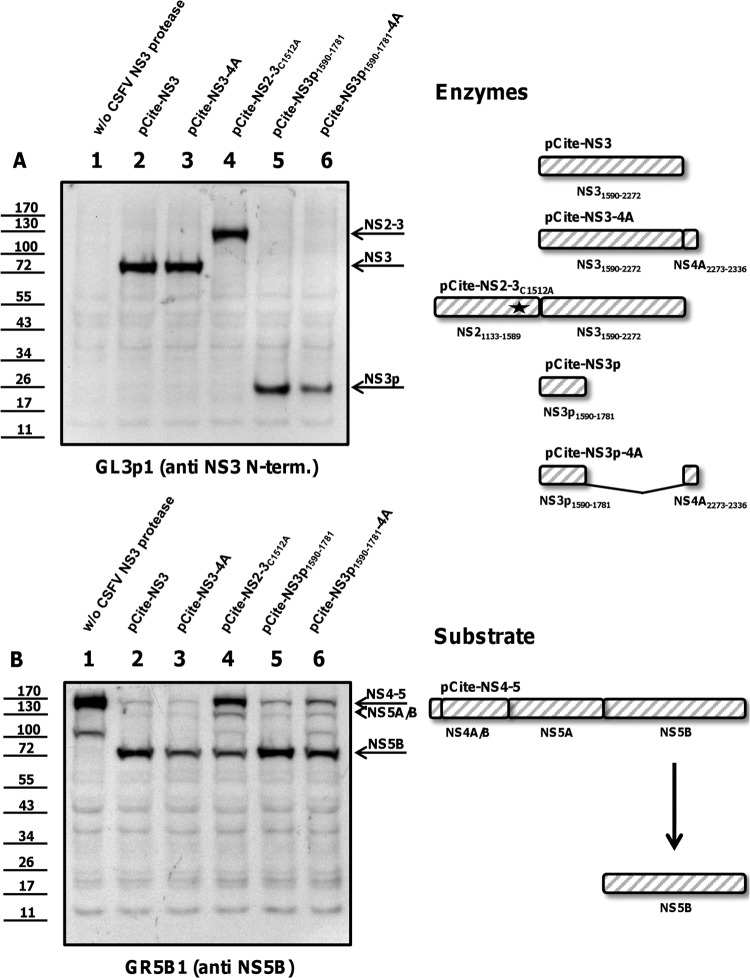
NS2-3 and NS3 are proteolytically active. BHK-21 cells were infected with vaccinia virus MVA/T7 pol, infected with MVA/T7 pol, and transfected with the NS4-5 substrate alone or cotransfected with plasmids encoding NS3 protease constructs (NS2-3, NS3) together with the substrate. (A) Expression of the enzymes (NS2-3, NS3) is documented by Western blotting using the moAb GL3p1. (B) Enzymatic activity is analyzed with a moAb against NS5B (GR5B1). The positions of NS2-3, NS3, NS4-5, and mature NS5B are indicated at the right. A scheme of NS2-3 and NS3 protease, NS4-5 substrate, and NS5B product is shown at the right. Molecular mass standards are shown on the left.
As we suspected that NS3p does not accumulate in the course of CSFV infection due to degradation, proteasome inhibitors were applied. At 18 h after infecting SK6 cells with ncp and cp CSFV at an MOI of 1, two different proteasome inhibitors (MG-132 and bortezomib) were added to the cells. After 6 h, cells were lysed and analyzed by Western blotting. Interestingly, no protein corresponding to NS3p (26 kDa) was identified on the blots using the GL3p1 moAb. Instead, the antibody recognized a novel protein species of 78 kDa after proteasome inhibitor treatment. The 78-kDa protein very likely represents the uncleaved fusion protein of NS2 and NS3p (NS2-3p) (Fig. 2D). Taken together, the results shown that internal cleavage within CSFV NS3 as well as NS2-3 results in a separation of protease and helicase domains at so-far-unknown cleavage sites.
Internal cleavage of NS3 is autocatalytic.
To further study the observed cleavage event, a bacterial expression system was established. Analogous to the proteolytically active single-chain HCV NS3-4A constructs (11), a single-chain CSFV NS4A3 protease was designed (Fig. 3A). For cofactor activity of the BVDV NS3 protease, a minimal essential NS4A peptide has been determined for amino acids 2293 to 2329 (4). The corresponding peptide-encoding sequence was fused via a GlySerGlySer (GSGS) coding linker to the NS3 gene (aa 1590 to 2272) and cloned into modified prokaryotic expression vector pET11a, providing a polyhistidine tag. NS4A3 was expressed in E. coli and purified by IMAC. Immediately after purification, the expected 90-kDa protein was obtained (NS4A3; Fig. 3B, 0 h), together with a copurified protein fragment of 35 kDa containing the His tag (data not shown). Within an incubation period of 2 h at 37°C or long-term storage at 4°C (not shown), a shift to protein bands of about 75 kDa, 58 kDa, 55 kDa, 35 kDa, and 14 kDa was observed (Fig. 3B, 2 h). In Western blot analysis, moAb GL3h1 against the C terminus of NS3 recognized the 90-kDa, 75-kDa, 58-kDa, and 55-kDa bands. MoAb GL3p1 against the N terminus of NS3 specifically reacted with protein species of 90 kDa, 75 kDa, and 35 kDa (Fig. 3C). The 90-kDa protein represents the expression construct NS4A3. The 75-kDa band represents an NS3 molecule containing the epitopes of both NS3-specific antibodies and lacking the 7His-TEV-NS4A polypeptide. In contrast to NS4A3, the 75-kDa protein shows no reactivity with the anti-NS4A antibody (data not shown). The bands of 55 and 58 kDa were exclusively detected by moAb GL3h1 and represent dominant C-terminal products of an internal NS3 cleavage (C-term. NS3; Fig. 3B and C, 2 h). The generation of the 55-kDa fragment occurring after autoproteolysis of recombinant NS4A3 might resemble processes in CSFV-infected cells. The corresponding 58-kDa fragment, which is less abundant in infected cells, might be further processed into the stable 55-kDa product. The 35-kDa band was detected by moAb GL3p1 only and likely represents 7His-TEV-NS4A-NS3p. The N-terminal cleavage product of 35 kDa was unstable, showing weak signals after 2 h of incubation. A 14-kDa protein band, which is generated only after incubation, is very likely the stable N-terminal product of NS4A3 but is no longer recognized by the protease-specific antibody.
The breakdown of the bacterially expressed NS4A3 was dependent on the presence of the NS3 protease and the NS4A cofactor. Substitution of active site Ser1751Ala (NS4A3S1751A) or deletion of essential parts of the NS4A cofactor (ΔSerLysArgHisIleProValValThr, NS4Adel3) yielded a stable product of 90 kDa (Fig. 3D).
For determination of the locations of intramolecular cleavage sites, the N termini of both helicase fragments (Fig. 3B, C-term. NS3) were analyzed by protein sequencing. Microchemical analyses revealed the N-terminal KGWSGL sequence for the 58-kDa fragment and the MSGIQT for the 55-kDa fragment. This corresponds to cleavage events at the L/K peptide bond at positions 1748 and 1749 and the L/M peptide bond at positions 1781 and 1782 (Fig. 4). A leucine residue is conserved at the P1′ position of all known NS3 cleavage sites (cleavage site position nomenclature according to reference 29). Lysine or methionine residues at the P1 position have not been reported for the pestivirus NS3 protease.
Fig 4.
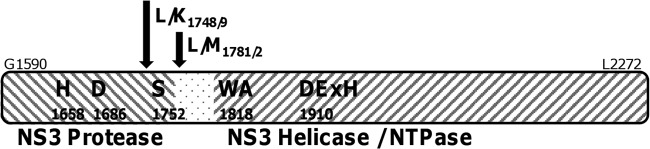
Organization of pestiviral NS3. CSFV NS3 extends from residue G1590 to L2272. Active side residues of the serine protease moiety include H1658, D1686, and S1752. Characteristic motives of the helicase start at residue 1818 with Walker motive A. The active-side DEYH box starts with D1910. Internal NS3 cleavages were mapped to residues L/K1748/9 and L/M1781/2.
The larger protease fragment is proteolytically active.
The cleavage at Leu1748 occurs N-terminally of Ser1752, an active site residue of the catalytic triad His/Asp/Ser (Fig. 4). NS3 protease fragments lacking Ser1752 are proteolytically inactive, as shown in earlier studies on the NS3 protease of BVDV (4). To assess the proteolytic activity of the larger fragment extending to Leu1781, a trans-cleavage assay as described by Tautz et al. (4) was adapted to CSFV nonstructural proteins. In transient expression in BHK cells using vaccinia virus MVA/T7 pol, plasmids encoding different NS3 enzymes (detected with moAb GL3p1 as shown in Fig. 5A, 6A, and 7A) were cotransfected with a plasmid encoding the cleavable substrate NS4-5 (detected with moAb GRS5b1 as shown in Fig. 5B, 6B, and 7B). Corresponding experiments share the same number labeled on top of each lane; thus, Fig. 5A, lane 1, and Fig. 5B, lane 1, show one experiment analyzed with two different antibodies. The experimental setup was controlled using a vaccinia virus mock transfection (Fig. 5, lanes 1) and transfection of the substrate alone (without [w/o] CSFV protease; Fig. 5, lanes 2) and of the substrate together with enzymes NS2-3 (Fig. 5, lanes 3) and NS3 (Fig. 5, lanes 4). The uncleaved NS4-5 substrate is visible in the absence of a CSFV NS3 protease (Fig. 5B, lane 2). NS3 and NS2-3 facilitated trans-cleavage to release mature NS5B (Fig. 5B, lanes 3 and 4). Self-cleavage of wild-type NS2-3 was not detected after expression in BHK cells, as shown earlier for BVDV NS2-3 by Lackner et al. (25). To further secure the finding that the uncleaved NS2-3 precursor acts in trans on NS4-5 substrates, the substitution Cys1521Ala was introduced. This mutation inactivates the cryptic NS2 protease abrogating NS2-3 cleavage (Fig. 6A, lane 4). The uncleavable NS2-3 was also capable of processing the NS4-5 precursor to mature NS5B, though the efficiencies of NS2-3 proteases were markedly reduced compared to those of mature NS3 (Fig. 5B, lane 3, Fig. 6B, lane 4, and Fig. 6B, lane 2).
Fig 6.
NS3p is proteolytically active. BHK-21 cells were infected with vaccinia virus MVA/T7 pol and transfected with the NS4-5 substrate or cotransfected with plasmids encoding enzymes and substrates. (A) Expression of the enzymes (NS3, NS3-4A, NS2-3C1512A, NS3p, and NS3p-4A) is documented by Western blotting using moAb GL3p1. Autocatalytic NS3-4A cleavage removes the 4A moiety of NS3-4A and NS3p-4A. Introduction of C1512A abrogates the activity of autocatalytic NS2 protease, to prevent NS2-3 cleavage. (B) Enzymatic activity of the constructs was analyzed with a moAb GR5B1. The positions of NS2-3, NS3, NS3p, NS4-5, and mature NS5B are indicated at the right. A scheme of NS3 enzyme constructs, NS4-5 substrate, and NS5B product is shown at the right. Molecular mass standards are shown on the left.
Fig 7.
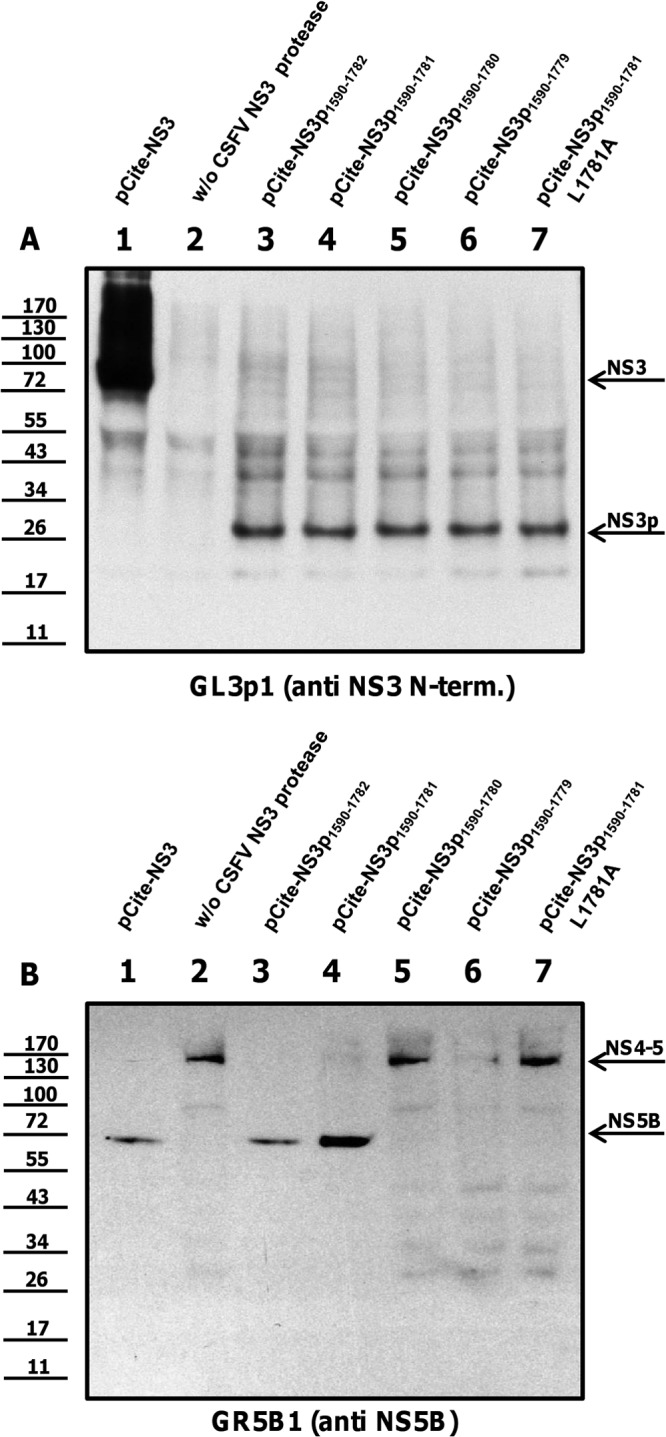
Leu1781 is the C-terminal border of the NS3 protease. BHK-21 cells were infected with vaccinia virus MVA/T7 pol and cotransfected with NS3 and NS4-5 constructs (positive control). BHK-21 cells were infected with vaccinia virus MVA/T7 pol and transfected solely with the substrate construct plasmid (negative control) or cotransfected with truncated NS3 enzymes and substrates. (A) Expression of enzymes is documented using moAb GL3p1. (B) Enzymatic activity of NS3 fragments is analyzed with moAb GR5B1. The positions of NS3, NS3 fragments, NS4-5, and mature NS5B are indicated at the right. Molecular mass standards are shown on the left.
We analyzed the cis-cleavage activity of NS3p1590–1781 with the transfection of an NS3p-4A fusion protein preserving the authentic L/S cleavage site. Under these conditions, NS4A is removed from the C terminus of NS3p-4A and from the C terminus of the positive control NS3-4A. Thus, only the 26-kDa NS3p and the 75-kDa NS3 are visible after expression of NS3p-4A and NS3-4A (Fig. 6A, lanes 2 and 3).
Systematic C-terminal truncation experiments revealed that NS3p1590–1781 is the shortest protease molecule with proteolytic activity (Fig. 7, lanes 4). Removal of leucine 1781 (for example, NS3p1590–1780) abrogated proteolytic activity (Fig. 7, lanes 5). Furthermore, not only deletion but also an exchange of Leu1781Ala (NS3p1590–1781L1781A) abolished the protease activity of the protease (Fig. 7, lanes 7).
Relevance of internal NS3 cleavage site residues for virus growth.
Sequence alignments of pestiviruses revealed that both internal NS3 cleavage sites are well conserved among the different pestivirus species. While Leu1748 is invariable, the leucine at position 1781 is an isoleucine residue in many BVDV strains (e.g., BVDV-1, strain Oregon C24V; AAC61755.1) and in one instance (BVDV-1 799cp; BAD04933.2) a phenylalanine residue is reported. Even the newly discovered Bungowannah virus, which is distantly related to CSFV, matches both leucine residues. A series of mutations of leucine residues 1748 and 1781 was introduced in the CSFV replicon. Most of these replicon RNAs (p447repΔL1748, p447repΔL1781, p447repL1748A, Leu1781A, and Leu1781G) were nonreplicative. The mutants carrying Leu1748G, Leu1748N, and Leu1781N yielded a very small number of cells that showed antigen expression, as assessed by NS3 detection (data not shown). In contrast, in more than 80% of the cells transfected with wild-type CSFV replicon RNA, the NS3 antigen was detectable. Because replication levels were too low and revertants could not be selected, the replicon system was not appropriate for further studies.
As we were interested in the appearance of reversions, further analysis was done using full-length genomes that allowed rescue and selection of replicative genomes. The CSFV mutants with deletions of leucines (ΔL1748 or ΔL1781) were nonreplicative, and infectious virus was not rescued at any time, most likely because of a loss of protease function (data not shown). After transfection into SK6 cells, transcripts of p447L1748A, p447L1748G, and p447L1748N resulted in single foci of antigen-positive cells and progeny virus production was at least 400-fold reduced at 24 h posttransfection compared to wild-type CSFV production (p447; Fig. 8A). Two passages of the rescued viruses on fresh SK6 were done to perform sequence analyses. The rescued viruses revealed for position 1748 either reversion to the wild-type leucine or an arginine residue. In a direct comparison, the viability of p447L1748R was about 40-fold reduced compared to that of p447L1748 in terms of progeny virus production (Fig. 8A).
Fig 8.
Mutational analyses of NS3 residues L1748 and L1781. The mutations L1748A, L1748G, L1748N, L1748R, L1781A, L1781G, L1781N, L1781V, and L1781I were introduced in the CSFV clone Alfort/Tuebingen using reverse genetics. SK-6 cells were transfected with the synthetic genomic RNA by electroporation. Progeny virus was titrated from the supernatant 24 h and 48 h after transfection on confluent monolayers of SK-6 cells. Infection was allowed for 2 h before cells were covered with semisolid methyl-cellulose DMEM. At 24 h after titration, the cells were washed, fixed, and stained using an immunoperoxidase reaction and a moAb against CSFV NS3. Levels of infectious CSFV particles were determined as numbers of focus-forming units per ml (ffU/ml). (A) All tested mutations of NS3 residue L1748 resulted in at least 40-fold progeny virus titer reduction. (B) For L1781 solely, the exchange L1781I led to wild-type-like titers. Other amino acid exchanges of residue L1781 impaired viral growth more than 300-fold. Virus titers and error bars are calculated on the basis of the results of three independent experiments.
The substitution of Leu1781 by Ala (p447L1781A), Gly (p447L1781G), and Asn (p447L1781N) severely compromised viral growth. Viruses could be rescued after one passage of transfected cells that were further passaged on fresh cells until high-titer virus production was observed (2 to 3 passages; data not shown). Sequencing uncovered uniform reversions to Leu, Ile, or Val at position 1781. Introduction of an isoleucine 1781 codon into p447 resulted in viruses with wild-type-like growth, whereas p447L1781V displayed more than 300-fold-reduced viability compared to p447 (Fig. 8B) and rapidly acquired a further reversion to Leu or Ile (data not shown). These data suggest a strong functional selection for Leu at position 1748 and Leu/Ile at position 1781.
DISCUSSION
The pestivirus NS3 protease has been in the center of pestivirus research for almost 2 decades. Active site residues, cofactor requirements, substrates, and the complex biosynthesis characteristics have been elucidated in great detail (4, 30, 31). For this reason, it is more than a surprise to find novel autocatalytic, intramolecular cleavages that extend our knowledge about substrate dipeptides and the actual proteolytically active protease molecules. Mature NS3 has been considered a protease that requires activation by NS2-3 cleavage, which is strictly regulated by the cellular cofactor JIV (17). It was further suggested that the zymogen/proenzyme NS2-3 is solely active in the NS4A cis-cleavage and unable to mediate the trans-cleavages between NS4A, NS4B, NS5A, and NS5B. Our studies reveal that uncleaved NS2-3 is capable of breaking down NS4-5 precursors in trans-cleavage assays without any activation step. As there is no reason to exclude the possibility that this activity occurs also during CSFV infection, the current concept of regulating the polyprotein processing and replication needs to be modified. In addition to this, the internal cleavage site at Leu1781/Met1782 releases a fully active protease that is no longer fused with the helicase domain. While the first evidence for this intramolecular cleavage—the 55-kDa variant of NS3—was observed but not pursued in the past, the availability of serological reagents specifically recognizing N- or C-terminal NS3 peptides provided new details. This C-terminal fragment that comprises the entire helicase domain is detectable in almost equal concentrations in cells infected with ncp, cp CSFV, or CSFV subgenomes. In contrast, the N-terminal 26-kDa fragment consisting of the protease was apparent only in cells infected with rapidly replicating CSFV subgenomes. The same applies to the smaller 24-kDa NS3p fragment that derives from the Leu1748/Lys1749 cleavage. Blocking proteasomal degradation in ncp and cp CSFV-infected cells (18) led to a surprising result. Instead of the expected 24-to-26-kDa protein, a novel 78-kDa protein species was recognized by the NS3p-specific antibody. This molecular mass fits with a fusion protein that encompassed NS2 and NS3p occurring after intramolecular cleavage within the NS3 moiety of NS2-3. The intensities of 78-kDa bands differ between ncp and cp CSFV. It can be explained by the more efficient cleavage of NS2-3 by the NS2 autoprotease in cp CSFV-infected cells lowering the levels of NS2-3 precursors (Fig. 2D). Nevertheless, the resulting NS3p is not sufficiently stabilized by proteasome inhibitors to become detectable in cp CSFV-infected cells.
We have tried to address the issue of whether the novel intramolecular cleavage of NS3 is important for CSFV replication. Deletion of Leu1781 resulted in inactive protease and nonreplicative RNA. Substitution of Leu1781 with other residues, with the exception of isoleucine, was not tolerated (Fig. 8B). Hence, all other studied residues reverted quickly to leucine or isoleucine residues in the context of replicative CSFV systems. Leu/Ile1781 is of key importance for the NS3 protease activity (Fig. 7, lanes 7). It can be safely assumed that Leu1781 is essential for catalysis, and we therefore believe that not the internal cleavage at this residue but the sequence identity of the residue is pivotal for CSFV replication. Structural data of pestivirus NS3 that could be helpful to further address this issue are still missing. According to our data, the minimal active protease fragment consists of 190 amino acids (1592 to 1781) and is thus 19 amino acids smaller than that previously reported (209 amino acids [1592 to 1800]) (4). In comparison, the minimal essential protease domain of HCV was mapped to 178 aa (8, 12) and the yellow fever virus NS3 protease to 180 aa (32). As shown in Fig. 9, the C-terminal internal NS3 cleavage site locates in a C-terminal protease stretch which is not conserved between the genera pestivirus and hepacivirus. This interdomain linker sequence was previously suggested to be involved in regulation of Flavivirus replication (33).
Fig 9.

Multiple sequence alignment of NS3 protease/helicase junction. The protein sequences of CSFV (strain Alfort; Protein Information Resource [PIR] accession no. GNWVHC), BVDV-1 (strain NADL; GenBank NP_776267), dengue virus 1 (DEV-1) (strain Brazil/97-11/1997; NP_722463.1), yellow fever virus (YFV) (isolate Ivory Coast/85-82H/1982; Q98803), hepatitis C virus subtype 1a (HCV1a) (isolate 136D_3048_4428_4; ADV92090), and HCV1b (strain MD4-1; AF165051.1) were aligned using clustal omega (version 1.1.0). Positions of the active site serine of the protease domain (vertical arrow) and the Walker A motive of the helicase (horizontal arrow) are indicated. The internal sites of NS3 autocleavages are depicted by inverted triangles. The minimal active protease domain for each virus is marked in bold. Identical, homologous, and similar amino acid residues are indicated below the alignment.
What is the relevance of the intramolecular NS3 cleavages for virus biology? We currently cannot answer this question, but the cleavage is certainly no coincidence. The unusual dipeptide Leu/Met indicates the presence of a suboptimal substrate; thus, the cleavage is not as efficient as it is at the other sites. Supporting this hypothesis, the “improvement” of the Leu/Met site to an Leu/Ser site in the infectious cDNA resulted in massively impaired virus replication with >2,000-fold-reduced viability (data not shown) and reversion, indicating a need for low-level cleavage. It can be further speculated that internal NS3 cleavage is beneficial, probably because the different protease- and/or helicase-containing molecules (NS2-3, NS2-3p, NS3, NS3p, NS3h) might serve other functions.
Considering the release of minute amounts of a soluble and probably freely diffusing chymotrypsin-like protease in the cytosol of an infected cell, the targeting of viral or host proteins might be involved in innate immunity or apototic transduction cascades. Such effects may be dangerous for the virus, at least for those pestiviruses that perpetuate persistent infections. The short life span of NS2-3p combined with the autocatalytic cleavage at leucine 1748 may therefore represent a safety measure. As identified by N-terminal sequencing, the helicase domain of NS3 is undamaged with regard to the integrity of Walker motives. In-depth functional analyses of the helicase function of the fragments left by internal cleavage are necessary before we can speculate about a putative function of a protease-free helicase/NTPase. This also applies to the recent finding that the helicase domain is strongly involved in virus particle formation (19). In similarity to four protease-containing molecules (NS2-3, NS2-3p, NS3, and NS3p), there are three helicase-containing moieties: NS2-3, NS3, and NS3h. NS3h is probably innocuous as it accumulates in the infected cell. One possible function of NS3h could be envisioned as that of a dominant-negative factor regulating replication complex formation. Internal processing of NS3 has also been reported for other members of Flaviviridae, e.g., yellow fever virus, dengue virus (34, 35), and HCV (36). In either case, internal cleavages occur within the helicase domain of NS3 and yield C-terminally truncated molecules. While a single study suggested that cellular proteases mediate the internal NS3 cleavage in HCV (36), it was demonstrated that an autocatalytic protease activity of NS2B/NS3 or NS3/NS4A is responsible for internal NS3 processing (9, 13, 37, 38).
The autocatalytic cleavage of NS3 resulting in an active protease and a potentially active helicase likely resembles a primordial situation in the evolution of the unique flaviviral genome organization. Unrelated “protease” and “helicase” functions located initially to different polypeptides. Condensation of different enzymatic functions in a single protein follows strict size constraints, but a higher order of regulation may result. In this context, it is remarkable that the NS2-3 precursor which is considered to be essential for replication and particle formation is processed by two proteases. Different cofactor requirements, namely, (i) the cellular cofactor JIV for the NS2 autoprotease and (ii) viral NS4A for NS3, might determine the fate of NS2-3 and its products. Future experiments are necessary to determine the individual protein functions and whether or not the complex processing events of NS2-3 are interconnected.
ACKNOWLEDGMENTS
We thank Günther Lochnit (Institute of Biochemistry, Justus-Liebig-Universität Gießen) and MSD Animal Health (T.R.) for their support.
Footnotes
Published ahead of print 28 August 2013
REFERENCES
- 1.Simmonds P, Becher P, Collett M, Gould E, Heinz F, Meyers G, Monath T, Pletnev A, Rice C, Stiasny K, Thiel H-J, Weiner A, Bukh J. 2011. Family Flaviviridae, 1003–1020 In King A, et al.(ed), Virus taxonomy. Ninth report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA [Google Scholar]
- 2.Lindenbach B, Thiel H-J, Rice C. 2007. Flaviviridae: the viruses and their replication, p 1101–1152 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 3.Rümenapf T, Thiel H-J. 2008. Molecular biology of pestiviruses, p 39–96 In Mettenleiter T, Sobrino F. (ed), Animal viruses: molecular biology. Caister Academic Press, United Kingdom [Google Scholar]
- 4.Tautz N, Kaiser A, Thiel HJ. 2000. NS3 serine protease of bovine viral diarrhea virus: characterization of active site residues, NS4A cofactor domain, and protease-cofactor interactions. Virology 273:351–363 [DOI] [PubMed] [Google Scholar]
- 5.Warrener P, Collett MS. 1995. Pestivirus NS3 (p80) protein possesses RNA helicase activity. J. Virol. 69:1720–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamura JK, Warrener P, Collett MS. 1993. RNA-stimulated NTPase activity associated with the p80 protein of the pestivirus bovine viral diarrhea virus. Virology 193:1–10 [DOI] [PubMed] [Google Scholar]
- 7.Becher P, Orlich M, Thiel HJ. 1998. Complete genomic sequence of border disease virus, a pestivirus from sheep. J. Virol. 72:5165–5173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. 1994. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J. Virol. 68:5045–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falgout B, Pethel M, Zhang YM, Lai CJ. 1991. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J. Virol. 65:2467–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bazan JF, Fletterick RJ. 1989. Detection of a trypsin-like serine protease domain in flaviviruses and pestiviruses. Virology 171:637–639 [DOI] [PubMed] [Google Scholar]
- 11.Dimasi N, Pasquo A, Martin F, Di Marco S, Steinkuhler C, Cortese R, Sollazzo M. 1998. Engineering, characterization and phage display of hepatitis C virus NS3 protease and NS4A cofactor peptide as a single-chain protein. Protein Eng. 11:1257–1265 [DOI] [PubMed] [Google Scholar]
- 12.Taremi SS, Beyer B, Maher M, Yao N, Prosise W, Weber PC, Malcolm BA. 1998. Construction, expression, and characterization of a novel fully activated recombinant single-chain hepatitis C virus protease. Protein Sci. 7:2143–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kou YH, Chang MF, Wang YM, Hung TM, Chang SC. 2007. Differential requirements of NS4A for internal NS3 cleavage and polyprotein processing of hepatitis C virus. J. Virol. 81:7999–8008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agapov EV, Murray CL, Frolov I, Qu L, Myers TM, Rice CM. 2004. Uncleaved NS2-3 is required for production of infectious bovine viral diarrhea virus. J. Virol. 78:2414–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moulin HR, Seuberlich T, Bauhofer O, Bennett LC, Tratschin JD, Hofmann MA, Ruggli N. 2007. Nonstructural proteins NS2-3 and NS4A of classical swine fever virus: essential features for infectious particle formation. Virology 365:376–389 [DOI] [PubMed] [Google Scholar]
- 16.Lattwein E, Klemens O, Schwindt S, Becher P, Tautz N. 2012. Pestivirus virion morphogenesis in the absence of uncleaved nonstructural protein 2–3. J. Virol. 86:427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lackner T, Muller A, Pankraz A, Becher P, Thiel HJ, Gorbalenya AE, Tautz N. 2004. Temporal modulation of an autoprotease is crucial for replication and pathogenicity of an RNA virus. J. Virol. 78:10765–10775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamp B, Riedel C, Roman-Sosa G, Heimann M, Jacobi S, Becher P, Thiel HJ, Rumenapf T. 2011. Biosynthesis of classical swine fever virus nonstructural proteins. J. Virol. 85:3607–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riedel C, Lamp B, Heimann M, Konig M, Blome S, Moennig V, Schuttler C, Thiel HJ, Rumenapf T. 2012. The core protein of classical Swine Fever virus is dispensable for virus propagation in vitro. PLoS Pathog. 8:e1002598. 10.1371/journal.ppat.1002598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasza L, Shadduck JA, Christofinis GJ. 1972. Establishment, viral susceptibility and biological characteristics of a swine kidney cell line SK-6. Res. Vet. Sci. 13:46–51 [PubMed] [Google Scholar]
- 21.Stoker M, Macpherson I. 1964. Syrian hamster fibroblast cell line Bhk21 and its derivatives. Nature 203:1355–1357 [DOI] [PubMed] [Google Scholar]
- 22.Gallei A, Blome S, Gilgenbach S, Tautz N, Moennig V, Becher P. 2008. Cytopathogenicity of classical Swine Fever virus correlates with attenuation in the natural host. J. Virol. 82:9717–9729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riedel C, Lamp B, Heimann M, Rumenapf T. 2010. Characterization of essential domains and plasticity of the classical Swine Fever virus Core protein. J. Virol. 84:11523–11531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sutter G, Ohlmann M, Erfle V. 1995. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 371:9–12 [DOI] [PubMed] [Google Scholar]
- 25.Lackner T, Thiel HJ, Tautz N. 2006. Dissection of a viral autoprotease elucidates a function of a cellular chaperone in proteolysis. Proc. Natl. Acad. Sci. U. S. A. 103:1510–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corapi WV, Donis RO, Dubovi EJ. 1988. Monoclonal antibody analyses of cytopathic and noncytopathic viruses from fatal bovine viral diarrhea virus infections. J. Virol. 62:2823–2827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Köhler G, Howe SC, Milstein C. 1976. Fusion between immunoglobulin-secreting and nonsecreting myeloma cell lines. Eur. J. Immunol. 6:292–295 [DOI] [PubMed] [Google Scholar]
- 28.Schägger H, von Jagow G. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368–379 [DOI] [PubMed] [Google Scholar]
- 29.Berger A, Schechter I. 1970. Mapping the active site of papain with the aid of peptide substrates and inhibitors. Philos. Trans. R. Soc. Lond. B Biol. Sci. 257:249–264 [DOI] [PubMed] [Google Scholar]
- 30.Tautz N, Elbers K, Stoll D, Meyers G, Thiel HJ. 1997. Serine protease of pestiviruses: determination of cleavage sites. J. Virol. 71:5415–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J, Mendez E, Caron PR, Lin C, Murcko MA, Collett MS, Rice CM. 1997. Bovine viral diarrhea virus NS3 serine proteinase: polyprotein cleavage sites, cofactor requirements, and molecular model of an enzyme essential for pestivirus replication. J. Virol. 71:5312–5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chambers TJ, Weir RC, Grakoui A, McCourt DW, Bazan JF, Fletterick RJ, Rice CM. 1990. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc. Natl. Acad. Sci. U. S. A. 87:8898–8902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo D, Wei N, Doan DN, Paradkar PN, Chong Y, Davidson AD, Kotaka M, Lescar J, Vasudevan SG. 2010. Flexibility between the protease and helicase domains of the dengue virus NS3 protein conferred by the linker region and its functional implications. J. Biol. Chem. 285:18817–18827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arias CF, Preugschat F, Strauss JH. 1993. Dengue 2 virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology 193:888–899 [DOI] [PubMed] [Google Scholar]
- 35.Teo KF, Wright PJ. 1997. Internal proteolysis of the NS3 protein specified by dengue virus 2. J. Gen. Virol. 78(Pt 2):337–341 [DOI] [PubMed] [Google Scholar]
- 36.Shoji I, Suzuki T, Sato M, Aizaki H, Chiba T, Matsuura Y, Miyamura T. 1999. Internal processing of hepatitis C virus NS3 protein. Virology 254:315–323 [DOI] [PubMed] [Google Scholar]
- 37.Bera AK, Kuhn RJ, Smith JL. 2007. Functional characterization of cis and trans activity of the Flavivirus NS2B-NS3 protease. J. Biol. Chem. 282:12883–12892 [DOI] [PubMed] [Google Scholar]
- 38.Yang SH, Lee CG, Song MK, Sung YC. 2000. Internal cleavage of hepatitis C virus NS3 protein is dependent on the activity of NS34A protease. Virology 268:132–140 [DOI] [PubMed] [Google Scholar]