Fig 3.
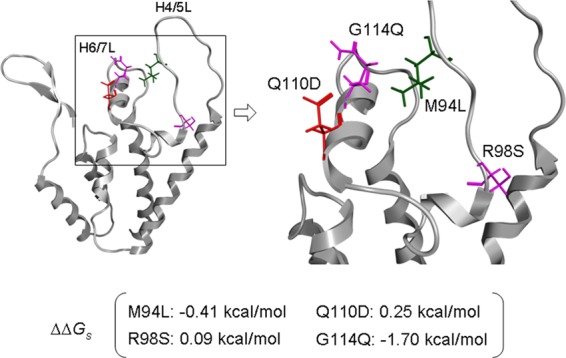
Structural analysis of the HIV-1mt CA NTD. A molecular model of the HIV-1mt CA NTD was constructed by homology modeling and refined as described previously (19). Single-point mutations were generated on the CA model, and ensembles of protein conformations were generated by using the LowMode MD module in MOE (Chemical Computing Group Inc., Quebec, Canada) to calculate average stability by using Boltzmann distribution. The stability scores (ΔΔGs) of the structures refined by energy minimization were obtained through the stability scoring function of the Protein Design application and are indicated below the structural model.
