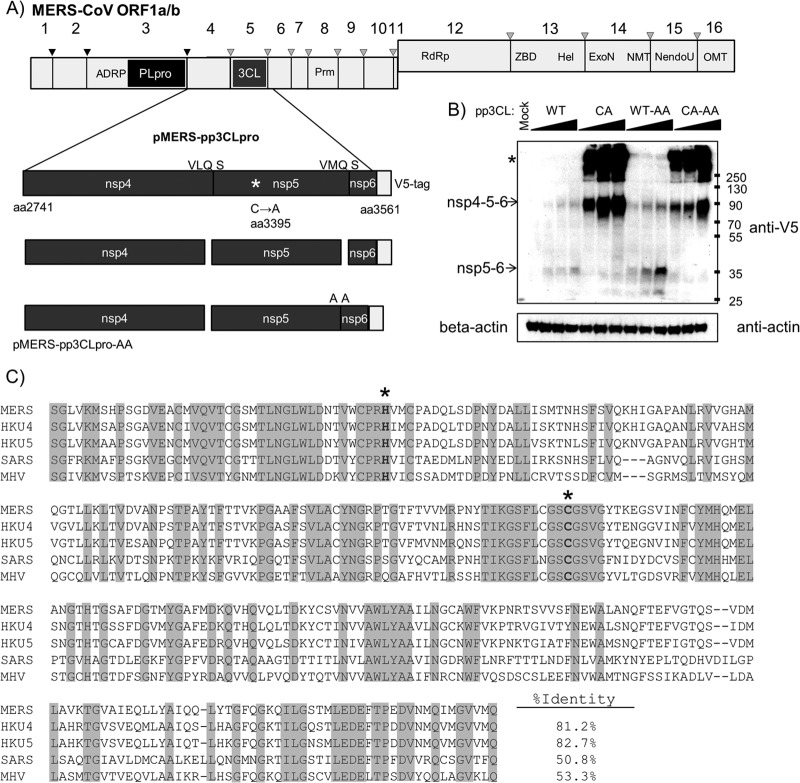
Fig 3.
MERS-CoV 3CLpro activity. (A) Schematic diagram of the 3CLpro domain and predicted cleavage sites in ORF1a/b. The region corresponding to the amino acid residues 2741 to 3561 spanning nsp4/5/6 was synthesized, cloned, and designated pMERS-pp3CLpro. The catalytic cysteine was changed to alanine, and the nsp5/6 cleavage site QS was changed to AA. (B) Expression and activity of 3CLpro. pMERS-pp3CLpro was transfected into HEK293T cells. Lysates were prepared at 20 h posttransfection and protein products analyzed by Western blotting. Anti-V5 detects polyprotein and processed products, and anti-beta-actin was used to monitor protein loading. *, protein aggregates. Numbers at the right are molecular masses (in kilodaltons). (C) Alignment of 3CLpro regions from selected betacoronaviruses. Alignments were performed using ViPR software MUSCLE alignment algorithms. Sequence identity is indicated with shading. Catalytic residues are boldface and marked with an asterisk. Accession numbers are listed in the legend to Fig. 1.