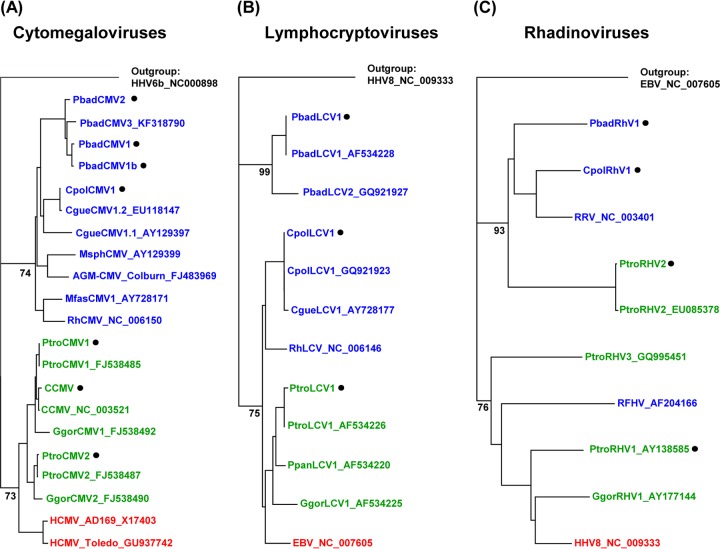
Fig 1.
Phylogenetic analysis. Partial gene sequences from the herpesviruses detected in this study and from published herpesviruses were aligned using MAFFT and subjected to phylogenetic construction of trees using the Geneious, version 5.5.7, tree builder (neighbor-joining module). Human, great ape, and OWM herpesviruses are shown in red, green, and blue, respectively. Viruses detected in this study are marked (black dot). (A) Tree based on glycoprotein B sequences of CMVs. (B) Tree based on DNA polymerase sequences of LCVs. (C) Tree based on gB sequences of RHVs. Bootstrap values are indicated at the basis of major clades and are suppressed at the tips of the clades.