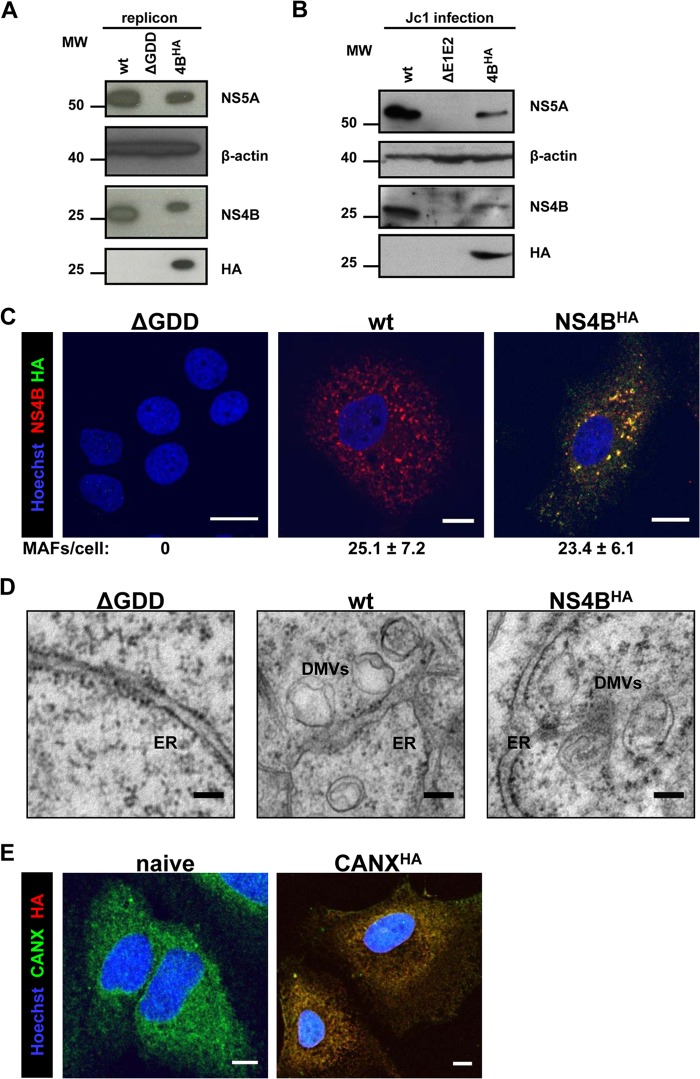
Fig 2.
Characterization of HA-tagged NS4B. (A) Huh7-Lunet cells were transfected with the in vitro-transcribed luciferase replicon RNAs specified at the top. Cells were harvested 72 h posttransfection and analyzed by immunoblotting, using the monospecific antibodies indicated on the right. The positions of molecular weight (MW; in thousands) markers are depicted on the left. (B) Huh7.5 cells were infected with the culture supernatants of cells transfected with the Jc1-derived full-length constructs specified at the top. After 72 h, cells were harvested and processed as described for panel A. (C) Huh7-Lunet cells were transfected with the in vitro-transcribed luciferase replicon RNAs specified at the top of each panel. After 48 h, cells were fixed, permeabilized with digitonin, and stained with NS4B- and HA-specific antibodies prior to confocal immunofluorescence microscopy. Only merged images are shown. Scale bars represent 5 μm. Numbers below each panel indicate the mean ± the standard deviation (SD) for NS4B-containing MAF per cell; for each condition, at least 10 different HCV-positive cells were analyzed. (D) Huh7-Lunet cells were transfected with in vitro-transcribed luciferase replicon RNAs as specified at the top of each panel. After 48 h, cells were fixed and flat embedded for TEM analysis. Scale bars represent 100 nm. (E) Naive Huh7-Lunet cells and those overexpressing CANXHA were fixed, permeabilized with Triton X-100, and stained with CANX- and HA-specific antibodies prior to confocal immunofluorescence microscopy. Only merged images are shown. Scale bars represent 5 μm.