Abstract
The emergence of the human 2009 pandemic H1N1 (H1N1pdm) virus from swine populations refocused public and scientific attention on swine as an important source of influenza A viruses bearing zoonotic potential. Widespread and year-round circulation of at least four stable lineages of porcine influenza viruses between 2009 and 2012 in a region of Germany with a high-density swine population is documented here. European avian influenza virus-derived H1N1 (H1N1av) viruses dominated the epidemiology, followed by human-derived subtypes H1N2 and H3N2. H1N1pdm viruses and, in particular, recently emerging reassortants between H1N1pdm and porcine HxN2 viruses (H1pdmN2) were detected in about 8% of cases. Further reassortants between these main lineages were diagnosed sporadically. Ongoing diversification both at the phylogenetic and at the antigenic level was evident for the H1N1av lineage and for some of its reassortants. The H1avN2 reassortant R1931/11 displayed conspicuously distinct genetic and antigenic features and was easily transmitted from pig to pig in an experimental infection. Continuing diverging evolution was also observed in the H1pdmN2 lineage. These viruses carry seven genome segments of the H1N1pdm virus, including a hemagglutinin gene that encodes a markedly antigenically altered protein. The zoonotic potential of this lineage remains to be determined. The results highlight the relevance of surveillance and control of porcine influenza virus infections. This is important for the health status of swine herds. In addition, a more exhaustive tracing of the formation, transmission, and spread of new reassortant influenza A viruses with unknown zoonotic potential is urgently required.
INTRODUCTION
Influenza A virus (IAV) infections cause economically important diseases in swine populations (1). In piglets and fattening pigs, benign respiratory forms of disease often prevail, yet they have a negative impact on the weight gain rates of affected animals. Influenza virus infections in sows may take a highly febrile course and have been considered a cause of prostaglandin-induced abortions and further fertility disorders. Control of swine influenza is difficult and depends on structural optimization of herd management as well as on the strategic use of highly efficacious vaccines (2, 3).
Until recently, in Germany, typical influenza-associated health problems affected swine herds mainly during the cold period of the year, resembling, although unlinked to, seasonal human influenza. Within recent years, porcine influenza rates have soared, especially in large industrial sow herds. Concomitantly, increasing rates of return to estrus and of abortions affecting sows and particularly gilts after service have been seen.
There is a close historic connection between human and porcine influenza viruses. Interestingly, viruses were usually seeded by the human population into swine herds. With the sole exception of the Asiatic influenza pandemic of the late 1950s (H2N2), all pandemic human IAVs found their way into swine populations. They adapted to the porcine host and continued to circulate independently of viruses in the human population while evolving into antigenically and phylogenetically discernible porcine lineages (4). Viruses of porcine lineages showed a lower tendency for antigenic drift than their counterparts in the human population (5). In addition to human sources, porcine influenza viruses also originated from avian precursor viruses: a subtype H1N1 virus lineage of purely avian origin established endemic infection in swine in Europe in the late 1970s. This lineage (H1N1av) dominated the epidemiology in European swine populations until very recently (5). Reassortants of this virus with human or human-derived lineages gave rise to H1N2 and H3N2 lineages in swine in Europe.
Serosurveys of swine have shown cocirculation of these three enzootic subtypes in different European countries at different prevalences (6). In addition, several reassortants between these lineages have been detected sporadically, but no further spread of any of these has been reported (7).
The most recent human pandemic influenza virus, of subtype H1N1, which emerged in 2009 (H1N1pdm virus) and harbors a multireassortant genome with several segments of porcine origin, is now also considered endemic in swine populations in several countries worldwide, including many European countries. It has already served as a new partner for reassortment with enzootic porcine lineages of subtypes H1N1av, H1N2, and H3N2 (8). In Germany, the H1N1pdm virus was first detected in a swine herd in December 2009. A reassortant virus carrying seven genome segments of the pandemic H1N1 virus and the neuraminidase (NA) of H1N1av (H1pdmN1) was reported in May 2010 (9). Beginning in early 2011, reassortants of subtype H1pdmN2 were found to circulate in five swine herds in Germany over a period of >1 year (10). The hemagglutinin (HA) sequences of these reassortants revealed unique characteristic coding mutations that set them apart from any other HA of pandemic/2009 origin.
Control of porcine influenza by vaccination is based on inactivated adjuvanted vaccines, although several studies have reported success with recombinant protein, DNA, and live-attenuated vaccines (2). The efficacy of an IAV vaccine depends on a close antigenic match between field and vaccine viruses and is challenged continuously by antigenic shift and drift of field viruses. In addition, interference from maternally derived antibodies can cause vaccination failure in piglets (11, 12). The antigenic drift of porcine influenza viruses in swine populations appears to be slower than that of influenza viruses in humans (13). This is probably due to the fact that the major part of the swine population is regularly removed by slaughter at the age of 6 to 8 months. Replacement with immunologically naïve piglets susceptible to antigenically “old” influenza virus strains allows for the continuing circulation of antigenically stable virus strains. The increasing rates of vaccination of sows observed in Germany in recent years have given rise to stronger immunological pressure, which eventually elicits accelerated antigenic drift of porcine IAV.
The current investigation was planned as a passive monitoring study for porcine influenza viruses in swine in the northwest of Germany. In the first phase, from 2009 to 2010 (study A), lung tissues and nasal swabs obtained from swine with severe respiratory disease and/or fertility problems were examined. In the second phase, between March 2011 and March 2012 (study B), nasal swabs from swine holdings with a record of IAV infections despite continuous vaccination of at least a part of the holding's swine population (usually the sows) were studied. Insight into the dynamic and ongoing viral evolution of swine influenza in an area with a high-density pig population was gained by characterizing virus subtypes with respect to their current phylogenetic and antigenic diversification. Experimental infections of swine with representative isolates of new reassortant lineages revealed virulent properties for their natural hosts.
MATERIALS AND METHODS
Study outline and sampling strategy. (i) Lung tissues.
The data of this study were established by molecular characterization of samples collected between 2009 and 2010 in northwest Germany, a region in which 55% of the German swine population (27 million pigs) is located. Sampling focused on pigs with a history of respiratory disorders in herds of 120 to 650 sows. Specimens of lung tissues obtained during postmortem examinations predominantly of weaners with clinical signs of respiratory disease were collected irrespective of further epidemiological stratifications.
(ii) Nasal swabs.
Between March 2011 and March 2012, a targeted sampling approach was followed for swine herds with pronounced respiratory and fertility problems. The study was mainly restricted to the same area as that for study A.
Selection criteria for herds were as follows: the herd size exceeded ≥200 sows, piglets were sold for fattening or were fattened on the farm, and the rate of return to estrus was higher than 15%; herds had been regularly vaccinated against influenza during the preceding 12 months; and there were problems with sows showing fever and reduced feed intake independent of the period of early lactation.
Selection criteria for animals within such herds comprised, for sows, a rectal temperature of >40°C independent of early lactation and reduced feed intake and, for weaners, clinical signs of acute respiratory disease.
Nasal swabs were to be taken by the consulting veterinarian from as many as 15 diseased sows and as many as 15 healthy sows in the same bay. In addition, or alternatively, nasal swabs were taken from as many as 20 acutely diseased weaners. A commercial swab system (Virocult; M&W), which was provided to the veterinarians together with a submission form, was used. With this form, consulting veterinarians were also asked to agree to give a telephone interview to further specify the clinical and epidemiological situation of the sampled herd. Samples were submitted without delay at ambient temperature for molecular and virological analysis.
Questionnaire.
Telephone interviews with consulting veterinarians who gave their consent on the submission form (exclusively for study B samples) were carried out on the basis of a pretested and validated questionnaire targeting numerous epidemiological parameters (available on request). Questionnaire data were recorded with InfoPath, and descriptive statistics were calculated in Excel. An English translation of the questionnaire is available in the supplemental material.
RNA extraction and detection of viral RNA.
RNA was extracted manually as described previously (10) using the Qiagen viral RNA kit. Alternatively, samples were extracted semiautomatically by use of the MagAttract Virus Mini Kit and a BioSprint 96 device (Qiagen). Lung sample homogenates were processed by use of the NucleoSpin 96 virus core kit (Macherey Nagel) and the Freedom Evo pipetting robot (Tecan) according to the recommendations of the manufacturer. Extracted RNA was then subjected to detection of a fragment of the M gene of the influenza virus RNA genome by real-time reverse transcription PCR (M1.2 RT-qPCR) using a method described by Fereidouni et al. (14). An “internal” control RNA (IC-2) was used to exclude false-negative results due to PCR inhibition (15). Quantification cycle (Cq) values of ≥40 were considered negative.
Subtype identification by partial sequence analysis.
Samples with Cq values of ≤33 in the M-gene-specific RT-qPCR were selected for subtype characterization of hemagglutinin (HA) and neuraminidase (NA) genes. Amplicons of the expected sizes obtained by conventional RT-PCRs using “Pan-HA” and “Pan-NA” primers as described by Gall et al. (16, 17) were cycle-sequenced, and sequences were subjected to a BLASTN2 search of public databases for subtype identification.
Virus isolation.
Samples with Cq values of ≤30 in the M-gene-specific RT-qPCR were selected for virus isolation in MDCK cell cultures grown in 6-well plates as described elsewhere (18). Supernatants of cultures showing cytopathic effects until 72 h of incubation at 35°C were subpassaged once in MDCK cells in 25-cm2 culture flasks. The hemagglutination titer in the supernatant was measured following a freeze-thaw cycle and clarification of debris. The HA and NA subtypes of the isolate were confirmed by sequencing as described above. Hemagglutination inhibition (HI) assays were not used for subtype identification.
Full-length sequencing and phylogenetic analysis of HA, NA, and “internal” genes.
Selected virus isolates or nasal swabs that were highly positive for influenza virus RNA in M-specific RT-qPCR (Cq values, ≤25) were used to determine and analyze full-length HA and, for a lesser number of samples, NA gene sequences (see Table S1A in the supplemental material). In general, primers reported by Hoffmann et al. (19) were used for full-length amplification of the HA, NP, NA, M, and NS genes with the One-Step SuperScript III amplification kit as recommended by the manufacturer (Invitrogen, Darmstadt, Germany). PB1, PB2, and PA gene segments were each amplified in two overlapping fragments using primers described by Li et al. (20). Amplification and sequencing were limited to the HA1 fragment of the HA gene (primer sequences available on request) when RNA from clinical samples had to be used. Sequences were assembled using the GCG software suite and were then submitted to the EpiFlu database (isolate identification numbers and accession numbers are listed in Table S1A and B in the supplemental material).
Phylogenetic analysis of HA and NA gene segments was based on alignments of the open reading frames of the HA1 fragment (nucleotides 1 to 1032, representing amino acids [aa] 1 to 344) and of full-length NA (1,401 nucleotides, representing 467 aa). Alignments of newly established sequences and selected database entries were calculated using the MAFFT algorithm (21) and were further optimized by manual editing using JalView (22). The Akaike criterion calculated by jModelTest 2 (23) was used to choose the most appropriate mutation models. Phylogenetic analyses consisted of a maximum likelihood approach (PhyML, accessed via the ACTG server [24]). The resulting tree topology was used to identify clusters and the most appropriate outgroup sequences for each cluster. The reordered alignments were then subjected to analysis in a Bayesian framework (MrBayes software suite) by following in principle the guidelines of Smith et al. (25). Midpoint-rooted trees were extracted and drawn using FigTree software (http://tree.bio.ed.ac.uk/software/figtree/). The graphics were further edited with Inkscape (http://inkscape.org/).
Experimental infection of swine.
Groups of three 13-week old minipigs each were experimentally infected with porcine influenza virus isolates R2035/11 (H1pdmN2) and R1931/11 (H1avN2). The animals were derived from a breeding herd at the Friedrich-Loeffler-Institut (FLI). All experiments had received legal approval from an ethics commission (LALLF M-V/TSD/7221.3-2.5-004/10). The pigs tested seronegative for influenza virus nucleoprotein (NP)-specific antibodies in a commercial blocking enzyme-linked immunosorbent assay (ELISA) (IDvet, Montpellier, France). Two pigs of each group were nasally infected with 106 tissue culture infective doses (TCID50) in 1 ml of cell culture supernatant. A nebulizer device (Wolfe Tory Medical, Salt Lake City, UT) was used for instillation. At day 1 postinfection (p.i.), a third pig was added as a sentinel of pig-to-pig transmission. Animals were observed clinically daily for 10 days, and rectal temperatures were recorded once daily. Nasal swab samples were taken at days 2, 4, 7, 10, and 14 p.i. and were examined by M-specific RT-qPCR. Blood samples were collected on days 0, 7, 10, 14, and 20 p.i. and were examined by NP-specific blocking ELISA and hemagglutination inhibition using a homologous antigen (see below). The animals were euthanized on day 24 p.i.
HI assay.
Selected virus isolates were used for antigenic characterization by HI assay. A panel of previously established postinfection sera from pigs (day 28 p.i.) and ferrets (day 21 p.i.) was used (10, 18). HI procedures followed the method described by Lange et al. (18), including treatment with bacterial neuraminidases. HI results were analyzed by hierarchical agglomerative clustering using the HCE3.5 software package (26, 27) to produce a gridded heat map of normalized titers and a 2-dimensional (2D) dendrogram representing antigenic relationships. Two-dimensional hierarchical clustering for rows and columns by the complete linkage method was based on a Euclidian distance matrix.
RESULTS
Incidence of porcine influenza virus infections in selected swine herds in northwest Germany.
In the frame of study A, 401 lung tissue specimens obtained mainly from weaners yielded 86 (21.4%) samples positive by M-specific RT-qPCR, of which 36 could be further subtyped, as shown in Table 1.
Table 1.
Subtype characterization of porcine influenza viruses from selected swine holdings in Northwest Germany, 2009 to 2012a
| Subtype | No. of samples in: |
Total no. (% of all viruses [% of fully subtyped viruses]) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study A (2009–2010) | Study B |
||||||||||||||
| Mo (2011) |
Mo (2012) |
||||||||||||||
| 3b | 4 | 5 | 6c | 7b | 8b | 9b | 10 | 11 | 12 | 1 | 2 | 3 | |||
| Fully subtyped viruses | |||||||||||||||
| H1N1 | 13 | 11 | 5 | 5 | 6 | 4 | 5 | 7 | 13 | 8 | 6 | 0 | 7 | 6 | 96 (38.6 [62.7]) |
| H1N2 | 7 | 3 | 1 | 0 | 2 | 2 | 4 | 3 | 0 | 1 | 1 | 0 | 1 | 2 | 27 (10.8 [17.6]) |
| H1N1pdm | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 (0.8 [1.3]) |
| H1pdmN2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 5 | 1 | 11 (4.4 [7.2]) |
| H3N2 | 6 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 2 | 1 | 1 | 3 | 1 | 17 (6.8 [11.1]) |
| Sum | 26 | 14 | 7 | 6 | 10 | 7 | 9 | 12 | 14 | 13 | 8 | 1 | 16 | 10 | 153 (61.4 [100.0]) |
| Partially subtyped and nonsubtyped viruses | |||||||||||||||
| H1Nx | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 (4.0) |
| HxN1 | 0 | 2 | 2 | 2 | 1 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 2 | 13 (5.2) |
| HxN2 | 0 | 0 | 0 | 1 | 1 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 (2.4) |
| HxNx | 50 | 4 | 3 | 1 | 0 | 1 | 0 | 0 | 1 | 2 | 0 | 2 | 0 | 3 | 67 (26.9) |
| Sum total | 86 | 20 | 12 | 10 | 12 | 13 | 10 | 12 | 15 | 15 | 9 | 3 | 17 | 15 | 249 (100.0) |
In study A, viruses were detected in the lung tissues of weaners with severe respiratory disease. Study B samples consisted of nasal swabs from sows, piglets, or weaners from large holdings with fertility problems. Each sample represents one holding. Double infections were found in five cases.
A double infection with H1N1 and H1N2 was detected; both viruses were counted.
A double infection with H1N1 and H3N2 was detected; both viruses were counted.
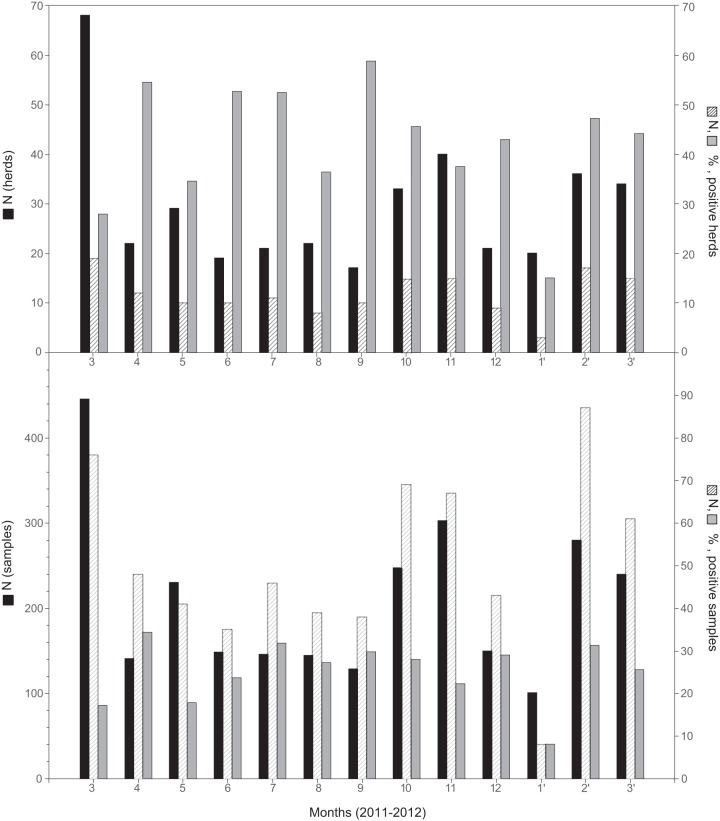
A total of 2,696 nasal swabs obtained from 382 pig herds originated from study B between March 2011 and March 2012; of these, 40.3% of holdings and 24.4% of samples tested positive for IAV RNA by M-specific RT-qPCR. Year-round virus circulation independent of the season was observed (Fig. 1).
Fig 1.
Frequency of porcine influenza virus infection in selected swine herds (top) and individual animals (bottom) sampled in northwest Germany, March 2011 to March 2012. Filled bars, absolute numbers of herds/animals sampled (left y axis); hatched bars, absolute numbers of herds/animals that tested positive (right y axis); shaded bars, percentage of herds/animals that tested positive (right y axis).
Most of the sample sets from one holding that tested positive contained at least one or two samples with Cq values of ≤33, a prerequisite for successful molecular subtyping or virus isolation, respectively. However, for roughly one-quarter of holdings with positive samples, no subtyping results were obtained due to small amounts and/or poor quality of viral RNA. A further 10%, particularly lung tissue samples, could be only partially subtyped (Table 1). Among the fully subtyped samples, H1N1 viruses of avian origin clearly dominated (62.7%), followed by subtypes H1N2 (17.6%) and H3N2 (11.1%). Pandemic H1N1 viruses were detected only on two holdings (1.3%), but reassortants between H1N1pdm and porcine HxN2 viruses (H1pdmN2) were found more frequently (7.2%). These reassortants emerged in 2011 and continued to circulate in 2012 (Table 1).
Questionnaire analysis (study B).
In the frame of the targeted study B, 236 submissions from sow-keeping farms were received, of which a total of 190 questionnaires could finally be included in an evaluation. The sample size is generally small, so that false-negative results cannot be firmly excluded. Since the majority of sample submissions were either from sows or from weaners, evaluation was separated as well (123 submissions for sows and 80 for weaners). Only 15 submissions contained samples from sows and weaners of the same holding. Few herds (n = 19) had been sampled more than once (17 were sampled twice, 1 three times, and 1 five times); these were ignored as repeated sampling and were rather treated as separate sample sets in the subsequent statistical description.
A total of 823 samples came from sows, of which 48 samples (5.8%) were influenza virus positive (13/123 herds [10.6%]). Samples for only two of the positive herds originated from older sows, and in four further positive herds, the age of the animals (gilts or older sows) was not specified. In seven cases, the positive samples were collected from gilts only. Subtyping revealed that most of these herds (n = 7) harbored H1N1av virus; H1N1pdm, H1N2, and H3N2 were each detected once; and in the remaining 3 herds, no complete subtyping was possible due to low concentrations of viral RNA.
Of 562 samples (representing 80 herds) from weaners, 181 (32.2%) were positive (43 herds [53.8%]). Again, H1N1av viruses made up the majority (17 herds), while H1N1pdm, H1N2, and H3N2 viruses were detected at lower frequencies (4, 5, and 4 herds, respectively). In one case each, simultaneous infections with H1N1av and H1N2 or with H1N1av and H3N2 were detected at the same time on a holding, but in different animals. Incomplete subtyping was recorded for 12 herds. The majority of positive samples originated from weaners aged 7 to 8 weeks.
Few factors analyzed via the questionnaire data appeared to be correlated (significance level, <0.05 by the F-test) with reduced risks of influenza virus infection (see Table S2 in the supplemental material). In sows, the most significant factor appeared to be vaccination against porcine reproductive and respiratory syndrome virus (PRRSV) (P = 0.002). Reduced frequencies of influenza virus infections were also seen in weaners that had received circovirus (porcine circovirus type 2 [PCV-2]) vaccination (P = 0.001) and in holdings that kept their diseased weaners and their piglets with retarded growth separated in sick bays (P = 0.007).
Phylogenetic diversity of porcine hemagglutinin (HA1) sequences from northwest Germany.
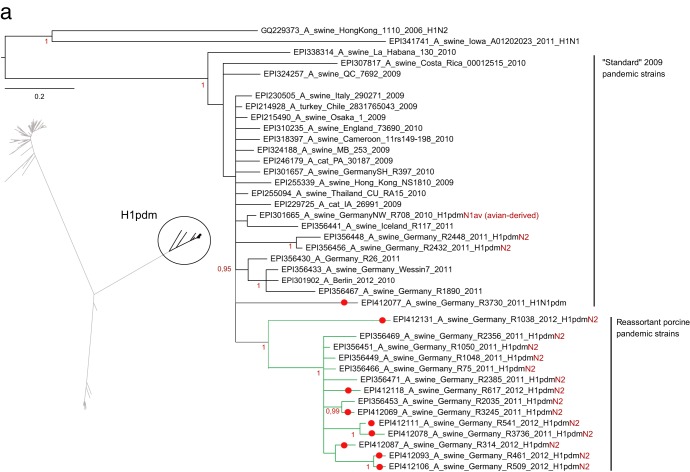
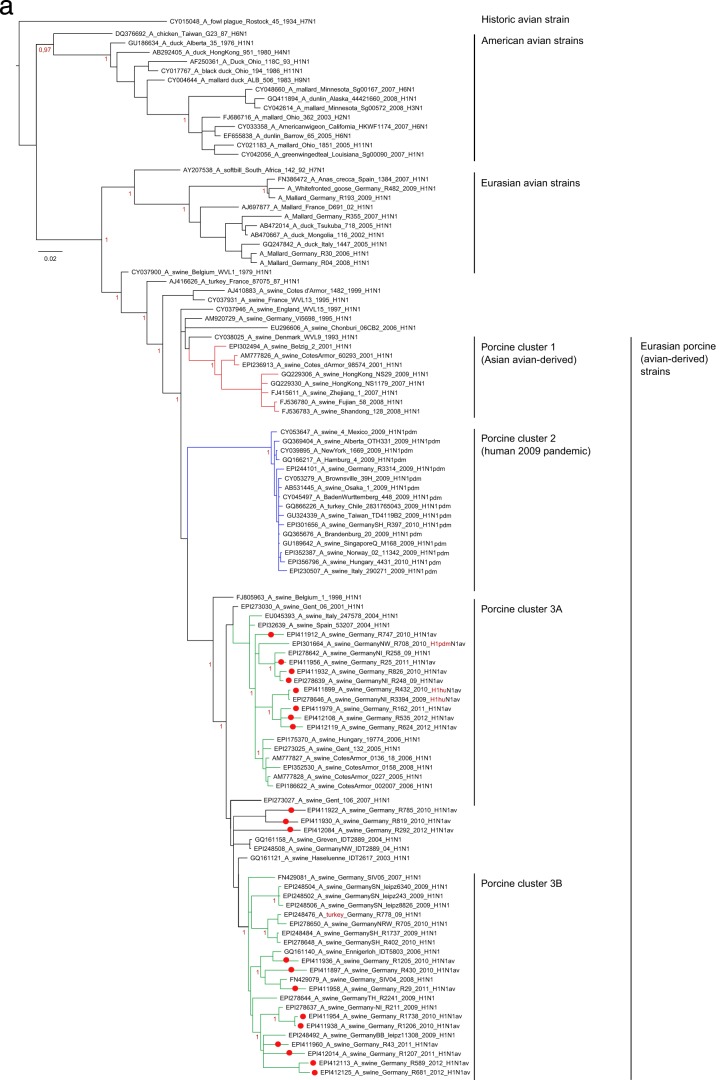
PhyML analysis confirmed the presence of three clearly distinct clusters within German porcine H1 sequences representing descendants of the human seasonal H1 virus (H1N2), the 2009 pandemic H1 virus (H1pdm), and the avian influenza virus-derived European porcine H1av virus (Fig. 2a to c, insets). Separate Bayesian analyses of each of these clusters unraveled distinct lineages within the H1pdm and H1av viruses cocirculating in swine in northwest Germany. A segregation into two separate lineages of the HA1 of H1pdmN2 reassortants within the cluster of H1pdm sequences (Fig. 2a) had been noticed earlier (10). This process continued in 2011 and 2012 (e.g., isolates R461/12 and R509/12 [Fig. 2a]). The HA1 sequences of these reassortant viruses were distinguished from “classical” pandemic H1 sequences by a set of six unique coding mutations [G172E, I183V, S200P, (S/T)202N, D204S, V338I].
Fig 2.
Phylogenetic analysis in a Bayesian framework of the hemagglutinin gene HA1 fragments of porcine influenza A viruses detected in selected swine herds in the northwest region of Germany, 2009 to 2011. (a) Subtype H1pdm; (b) subtype H1av; (c) subtype H1(N2); (d) subtype H3. Trees are drawn to scale as indicated by bars. Red dots on branches indicate sequences established in the frame of this study, for which specific registration numbers and years of isolation are given. EpiFlu database accession numbers can be retrieved from Table S1 in the supplemental material. Further sequences have been extracted from GenBank or the EpiFlu database, and their accession numbers are indicated in the trees. Clustering information is given by colored branches and/or by specific assignments to the right of the trees. Red color for subtype characterization indicates the presence of a reassorted neuraminidase segment. The screened trees in panels a to c represent a PhyML analysis of an enlarged set of porcine subtype H1 sequences to allow an overview of phylogenetic distances within this subtype. Details of the phylogenetic analysis are given in Materials and Methods.
More-complex clustering patterns were evident in the HA1 sequences of the porcine H1av viruses (Fig. 2b). At least four sublineages were distinguishable among recent H1av isolates from Germany. Two of these clusters were characterized by unique sets of coding mutations separating them from vaccine strain A/swine/IDT2617/2003 (H1N1), which has generally been used for vaccination on the holdings sampled in study B. Isolate R681/12, at the tip of cluster 1, accumulated 10 amino acid exchanges (N52T, S100P, N101D, A106T, K147R, V151I, S179K, I282V, H288R, E319K). Isolate R1856/11, at the tip of cluster 4, revealed substitutions V7I, T149S, A158I, G172K, L178I, K180T, and H288N (7 unique exchanges). Further groups of isolates, as well as single isolates, with fewer unique mutation patterns were noticed as well. This was found particularly for isolates positioned between clusters 1 and 2 (Fig. 2b), which show comparatively long horizontal branches (R369/09, R848/11, R1931/11, R3310/12). Two of these isolates also carry a reassortant NA of subtype N2.
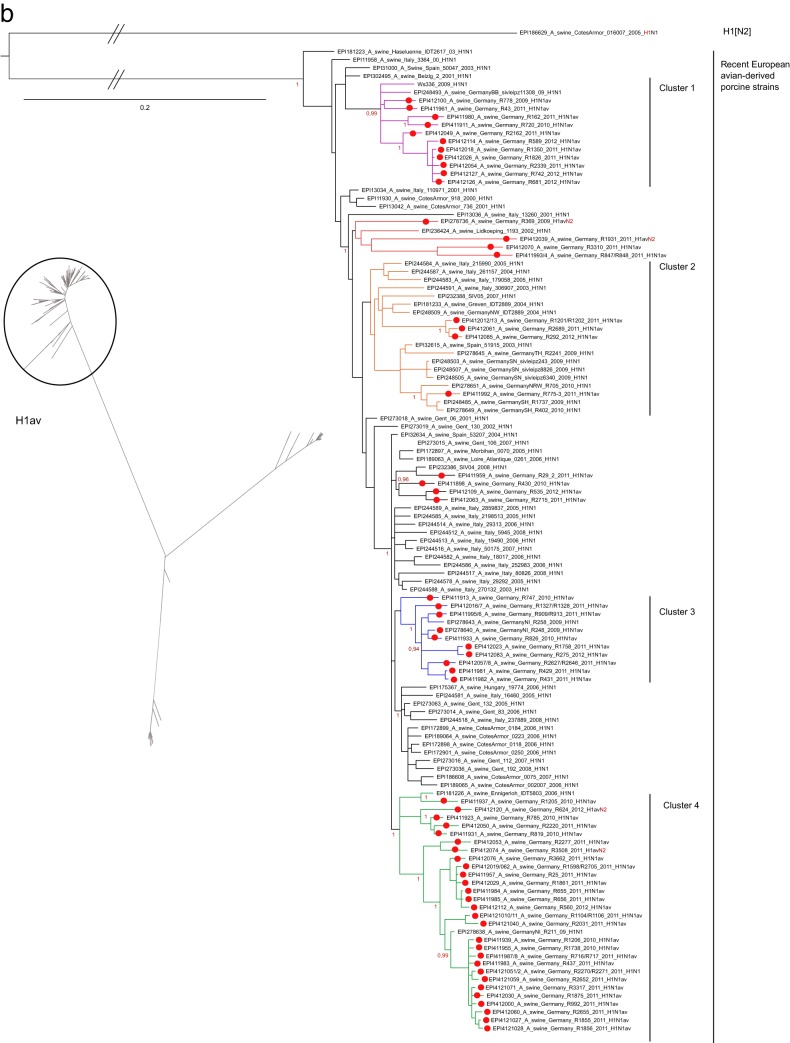
In contrast to those of the H1pdm and H1av lineages, HA1 sequences of the human-derived (hu) H1N2 porcine viruses recently detected in swine in Germany appeared to form a nearly homogenous phylogenetic group with only shallow furcations (Fig. 2c). Isolate R1421/10 was an exception and clustered outside this group. A PhyML analysis that included a wider selection of HA1 sequences of European H1N2 porcine isolates revealed a peculiar geographic restriction: contemporary viruses from France or the United Kingdom, Italy, and Germany formed distinct lineages, and R1421/10 clustered with older viruses from Central Europe (see Fig. S1 in the supplemental material). No such regional patterns were observed in PhyML analyses of H1av or H1pdm sequences from European swine (not shown).
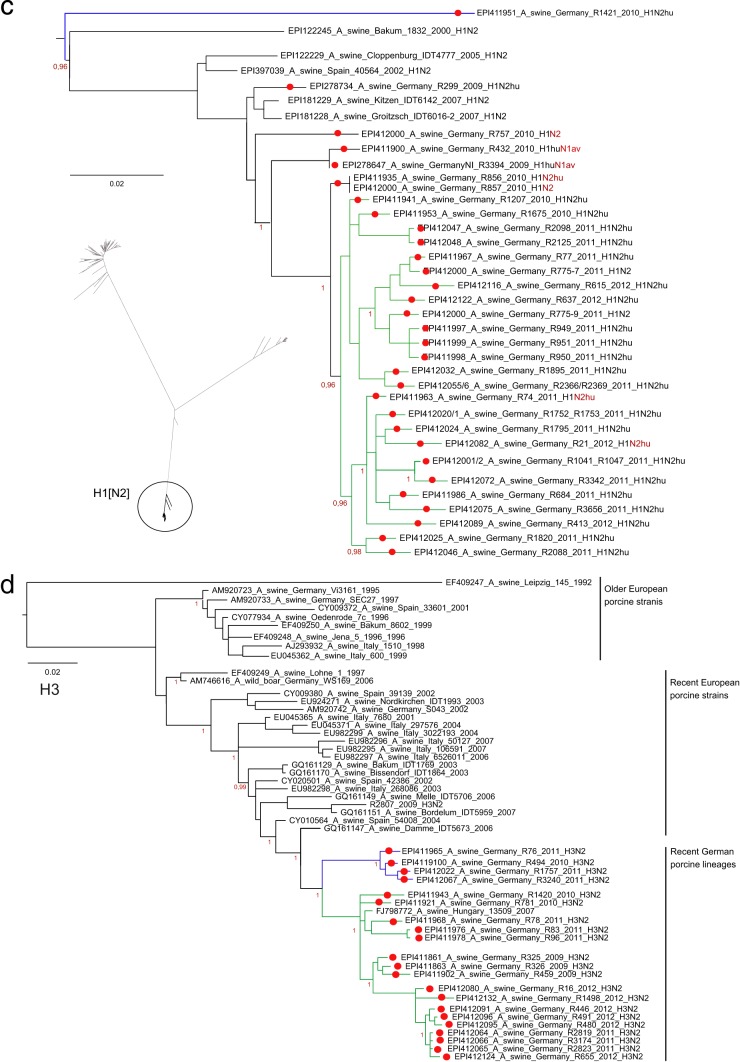
Recent porcine H3 HA1 sequences from Germany fell into a separate, slightly fissured cluster among European porcine H3 viruses (Fig. 2d). Unique coding mutations separated this lineage from older European porcine H3 isolates (N69K, K315R) and from each other (Fig. 2d, blue lineage) (V14G, G16S, D18S, D48N, L127I, V239I).
Phylogenetic diversity of porcine neuraminidase sequences from northwest Germany.
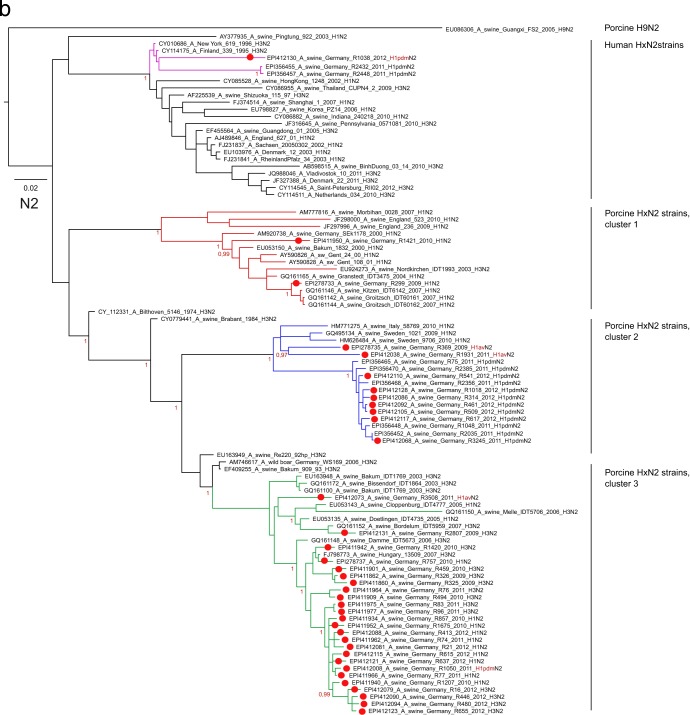
Clustering tendencies were also present in the NA phylogenetic trees. Porcine H1N1av N1 lineages (N1av) were clearly separated from truly avian lineages as well as from human seasonal and classical porcine lineages (Fig. 3a). The N1 of the human pandemic H1N1 viruses (N1pdm) formed a monophyletic group (cluster 2) within porcine N1av viruses (Fig. 3a). The N1av sequences of recent German porcine isolates formed two sublineages, but no correlation of these NA sublineages with the H1av clustering of the respective isolates was found.
Fig 3.
Phylogenetic analysis in a Bayesian framework of the neuraminidase genes of porcine influenza A viruses detected in selected swine herds in the northwest region of Germany, 2009 to 2011. (a) Subtype N1; (b) subtype N2. Trees are drawn to scale as indicated by bars. Red dots on branches indicate sequences established in the frame of this study, for which specific registration numbers and years of isolation are given. EpiFlu database accession numbers can be retrieved from Table S1 in the supplemental material. Further sequences have been extracted from GenBank or the EpiFlu database, and their accession numbers are indicated in the trees. Clustering information is given by colored branches and or by specific assignments to the right of the trees. Details of the phylogenetic analysis are given in Materials and Methods.
Deeper fissures were evident in the phylogenetic tree of N2 sequences (Fig. 3b), and recent German porcine isolates could be assigned to four clearly distinct clusters, three of porcine and one of human origin. Three of these clusters had already been delineated previously when the neuraminidase of H1pdmN2 reassortants was analyzed (10). The N2 sequences of the majority of nonreassorted H1N2 and H3N2 isolates grouped within porcine cluster 3 and were distinguished by the amino acid substitutions L23F, T236(I/V), I263V, and (L/I)360(T/I). Cluster 2 held most of the H1pdmN2 reassortant viruses, although two H1avN2 reassortants (R369/09, R1931/11) whose HA1 sequences were conspicuously distinct from all other H1av sequences also fell into this cluster. The N2 of porcine cluster 2 presented 12 unique amino exchanges [Y40C, (P/L)45S, N47(V/I), I57K, E74K, (K/R)75T, (P/T)126L, H197Y, F205L, (G/D/S)313N, (R/L)338Q, E343Q]. Only a few contemporary N2 porcine viruses from Germany segregated into porcine cluster 1. The N2 sequences of three further H1pdmN2 reassortants were assigned to the human HxN2 cluster and showed a close relationship to human N2 viruses that had last been detected in the human population in the mid-1990s. This N2 lineage has never been reported for European porcine influenza viruses; its origin remains uncertain.
Hemagglutinin and neuraminidase reassortant patterns and protein analysis.
Reassortants between the HA and NA of the different viral subtypes were found for the H1 subtype only. Reassortants between the H1N1pdm subtype and porcine viral neuraminidase subtypes N1 and N2 have been described recently (9, 10). Viruses of the H1pdmN2 lineages continued to be in circulation and evolved further in late 2011 and early 2012 (e.g., R509/12 [Fig. 3b]). In addition, reassortants between H1N2 HA and N2 derived from the porcine H3N2 lineage were observed: R757/10, R856/10, R74/11, and R21/12. Also, reassortants between the HA of the H1N1av-derived H1 lineage and NA N2 of different lineages were detected in a total of four cases in 2009 (R369/09), 2011 (R1931/11 and R3508/11), and 2012 (R624/12). One reassortant between H1N2 HA and H1N1av-derived NA N1 (N1av) was detected in 2009 (R3394/09).
Two reassortants, R369/09 and R1931/11, revealed HA protein sequences that were comparatively distant from the average H1av HA of contemporaneously circulating viruses, as signaled by long horizontal branches in Fig. 2b. R1931/11 showed deletion of a lysine residue at position 147. In addition, this HA sequence possessed a set of three additional predicted N-linked glycosylation sites (NGSs). Whereas all of the H1av HAs investigated share potential NGSs at amino acid positions (NetNGlyc 1.0 server scores) 28 (3+), 40 (3+), 498 (+), and 557 (2+), R1931/11 possessed further potential sites at positions 136 (NAT) (2+), 178 (NKS) (+), and 211 (NHT) (+).
The newly established neuraminidase sequences, whose accession numbers are listed in Table S1B in the supplemental material, were assessed (i) for mutations associated with decreased susceptibility, as confirmed by site-directed mutagenesis, to neuraminidase inhibitors oseltamivir (H275Y and N295S in N1; R292K, E119V, and N294S in N2) and zanamivir [Q136K and K150T in N1; R292K and E119(A/D) in N2] and (ii) for mutations observed during surveillance or following in vitro selection using oseltamivir (I223R in N1; I222V in N2). None of these resistance markers (28) were observed in the sequences investigated.
Genotyping.
Full-length genomes were established for 15 porcine isolates. For three additional isolates, one to two segments are missing (Table 2). Isolates were selected so as to represent a cross section of subtypes, HA/NA reassortment patterns, and years of isolation. PhyML analysis for “internal” segments (PB1, PB2, PA, NP, M, and NS) included the most closely related sequences from a BlastN2 search within the EpiFlu database as well as a random selection of additional sequences from Eurasian porcine and human isolates obtained since 2000. Genotyping results were obtained for each genome segment by use of the FluGenome tool (29) and are summarized in Table 2. The cassette of “internal” genome segments of Eurasian porcine IAV is composed of genotypes F, G, I, F, F, and 1E (for PB2, PB1, PA, NP, M, and NS) independently of the configuration of the HA and NA genome segments. Within each genotype, two sister clades were phylogenetically distinguishable and are tentatively referred to as “1” and “2” (see Fig. S2A to F in the supplemental material). This is consistent with the data for other German and European porcine IAVs of the past 10 years (sequences of 19 further viruses extracted from the EpiFlu database [data not shown]). Isolates R369/09 and R1931/11 exhibited PB2 sequence patterns, and R1931/11 also exhibited an NS sequence pattern, that could not be classified by the genotyping tool. This confirms the separate clustering of these isolates in the phylogenetic trees of the “internal segments” (see Fig. S2A to F in the supplemental material). The H1pdm isolates of porcine origin, including the H1pdmN2 reassortants (n = 3), belonged to genotypes C, D, E, A, F, and 1A (for PB2, PB1, PA, NP, M, and NS), a pattern similar to those of other H1pdm viruses isolated from pigs in Europe and elsewhere (Table 2). Based on these results, a total of three genome constellations, or genogroups (Table 2), were distinguishable.
Table 2.
Genotype assignments of porcine influenza viruses from swine holdings in Northwest Germany, 2009 to 2011
| Isolate | Genotype assignmenta |
Genogroupb | |||||||
|---|---|---|---|---|---|---|---|---|---|
| HA | NA | PB1 | PB2 | PA | NP | M | NSc | ||
| R248/2009 | 1av | 1av | G1 | F1 | I1 | F1 | F1 | 1E-1 | 1.1 |
| WS336/2009 | 1av | 1av | G1 | F1 | I1 | F1 | F1 | 1E-2 | 1 |
| R778/2009 | 1av | 1av | G1 | F1 | I1 | F1 | F1 | 1E-2 | 1 |
| R819/2010 | 1av | 1av | G1 | F1 | I1 | F1 | F1 | 1E-2 | 1 |
| R369/2009 | 1av | 2hu | G2 | ? | I2 | F2 | F2 | 1E-2 | 2.1 |
| R1931/2010 | 1av | 2hu | G2 | ? | I2 | F2 | F2 | ? | 2 |
| R299/2009 | 1hu | 2hu | G1 | F1 | I1 | F1 | F1 | 1E-2 | 1 |
| R3394/09 | 1hu | 1av | G1 | F1 | I1 | F1 | F1 | 1E-2 | 1 |
| R757/2010 | 1hu | 2hu | G1 | F1 | I1 | F1 | F1 | 1E-2 | 1 |
| R856/2010 | 1hu | 2hu | G1 | F1 | I1 | F1 | F1 | 1E-2 | 1 |
| R1421/10 | 1hu | 2hu | G1 | F1 | I1 | F1 | F1 | 1E-2 | 1 |
| R75/11 | 1pdm | 2hu | D | C | E | ND | ND | 1A | 3 |
| R2035/11 | 1pdm | 2hu | D | C | ND | A | F3 | 1A | 3 |
| R495/2012 | 1pdm | 2hu | D | C | E | A | F3 | 1A | 3 |
| R325/2009 | 3 | 2 | G1 | F1 | I1 | F1 | F1 | 1E-2 | 1 |
| R494/2010 | 3 | 2 | G1 | F1 | I1 | F1 | ND | 1E-2 | 1 |
| R2807/2009 | 3 | 2 | G1 | F1 | I1 | F1 | F1 | 1E-2 | 1 |
| R83/2011 | 3 | 2 | G1 | F1 | I1 | F1 | F1 | 1E-2 | 1 |
Genotypes were assigned according to the results obtained with the FluGenome genotyping tool and a series of PhyML analyses of the complete open reading frame of each of the internal genome segments (see Fig. S2A to F in the supplemental material). Numbers 1 and 2 specifying some of the genotypes refer to different sister clades within the genotype, as shown in the figures in the supplemental material. ND, no data available; ?, no unambiguous classification is possible by use of the FluGenome genotyping tool (25).
Summary of the genotype constellation of the internal segments.
All NS sequences clustered with allele A.
Characterization of “internal” gene segments.
The majority of the 15 non-H1pdm isolates analyzed here are, independently of their glycoprotein subtypes, true representatives of the Eurasian avian influenza virus-derived cassette of “internal segments.” Within this cassette, 19 unique amino acid exchanges that set these isolates apart from classical swine H1N1 viruses have been identified (30). Deviations from this pattern were evident in the NS segments of four isolates (R248/09, R819/10, R1931/11, and R535/12) [mutations W25Y, K66E, and (E/G)227R].
PB1 and PA segments encode further proteins in different reading frames. Differences in the lengths of the PB1-F2 proteins of European porcine influenza viruses have been identified (31). The majority of German isolates analyzed here (15 out of 18) encoded full-length (90-aa) PB1-F2. However, two isolates (R1737/09 and R2807/09) appeared to produce truncated forms of 79 aa and 87 aa, respectively. Two isolates (R369/09 and R1931/11) carried the rare N66S mutation in PB1-F2, which has been associated with an influence on virulence (32). The porcine H1pdm viruses from swine in Germany, like the human-derived H1pdm viruses, displayed a PB1-F2 peptide of only 11 aa. The PA-X protein-coding region, accessed via ribosomal frameshifting, could be demonstrated in all porcine PA sequences established here (Table 2). All but two isolates carried the most common decanucleotide motif at the proposed frameshift site (UCC UUU CGU C [33]). As analyzed by Shi et al. (34), the H1pdm viruses possess a stop codon at nucleotide 698, leading to a truncated PA-X protein (41 aa), whereas all other Eurasian porcine IAVs, including those analyzed here, should produce the full-length protein (61 aa).
Length differences were also observed in the NS1 protein. Full-length versions of 230 aa appear to be encoded by 11 out of 18 non-pdm swine influenza viruses (SIV), while the remaining 6 viruses for which NS sequences were established displayed truncated NS1 proteins of 217 aa, like some recent Italian isolates (35). These six truncated NS1 proteins were found in all three subtypes (twice each in H3N2, H1N2, and H1N1) and were isolated in 2009 and 2010. The length of NS1 was not correlated with HA/NA subtypes. The PDZ binding domain at the C terminus of full-length NS1 had the common sequence G-P-E-V (36) in the majority of isolates. In addition, R-P-E-V (R248/09) and R-P-K-V (R1931/11) occurred in one isolate each.
As expected from previous studies (37), the amantadine resistance markers S31N and R77Q in the M2 protein were observed in all the isolates analyzed here. A total of 16 non-pdm viruses also carried the L26I and V27(A/T) mutations in the M2 protein. V27(A/T) also confers amantadine resistance.
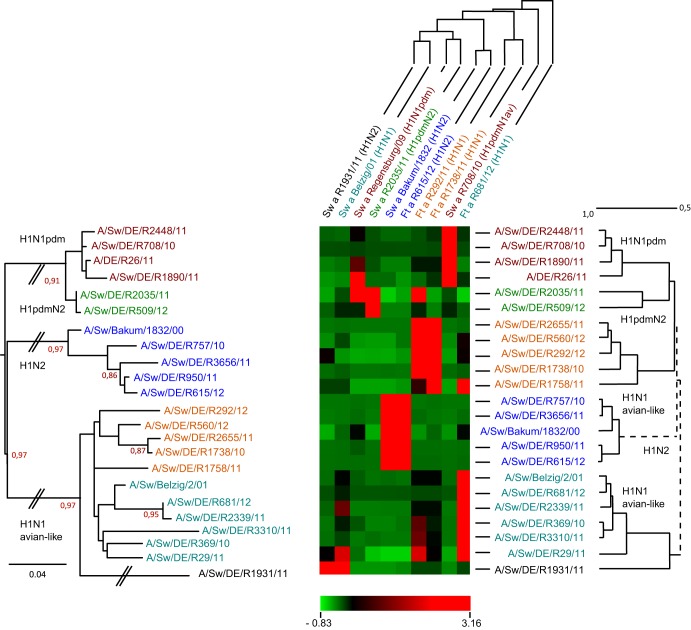
Antigenic patterns of selected porcine influenza A viruses.
Isolates representing the tips of the various sublineages discerned in the phylogenetic analyses were selected for antigenic analysis by HI assays using a set of postinfection sera established recently with swine and ferrets. An overview of the results for the antigenic relationships within subtype H1 is presented in Fig. 4. The titers obtained in the HI assays (see Table S3A in the supplemental material for absolute values) were transformed into a 2D antigenic map by agglomerative hierarchical clustering (HCE3.5). The heat map shown in Fig. 4 was set at a similarity of 0.50 (50%) for the rows. Within the larger clusters of H1pdm antigens, H1N2 and H1N1av lineages can be distinguished: (i) reassortant isolates of the H1pdmN2 subtype (R2035/11 and R509/12) (green) can be discerned from “true” H1pdm antigens (dark red); (ii) H1N2 viruses form a separate, homogenous cluster (dark blue); (iii) the H1av viruses are antigenically split into two clusters; and (iv) the reassortant H1avN2 isolate R1931/11 is distinguishable phylogenetically and, less pronouncedly, antigenically from other H1av isolates.
Fig 4.
Clustering of porcine influenza virus isolates of subtype H1. Two-dimensional antigenic hierarchical clustering based on HI titers was performed. The heat map shows three-color-coded normalized similarity values (light green, lowest similarity; black, intermediate similarity; bright red, highest similarity) (see the scale bar). The dendrogram to the right is drawn to scale according to the antigenic hierarchical clustering, and the viruses in this dendrogram each line up with a row in the heat map. The sera used in HI assays (listed above the heat map) have been given colors that match those of the viruses in the dendrogram to the right. The dendrogram to the left is based on phylogenetic analysis (PhyML) of amino acid sequences of the HA1 proteins of the same viruses used in cross-reactive HI assays.
Although the H3 viruses formed two clusters in the HA phylogenetic tree (Fig. 2d), a single homogenous group was found when these viruses were typed with serum raised against an older (Bakum/99) and a very recent (R655/12) H3N2 isolate (see Table S3B in the supplemental material). This finding is consistent with analyses of the commonly accepted antigenic sites within H3 HA1 (38), where only marginal changes were observed: D48N (antigenic site C) in 4 sequences, I212V (antigenic site D) in 2 sequences, and I276T (antigenic site C) in 3 out of the 4 sequences of the small blue cluster in Fig. 2d. No differences were seen with regard to potential N-glycosylation sites (n = 8).
Experimental animal infections.
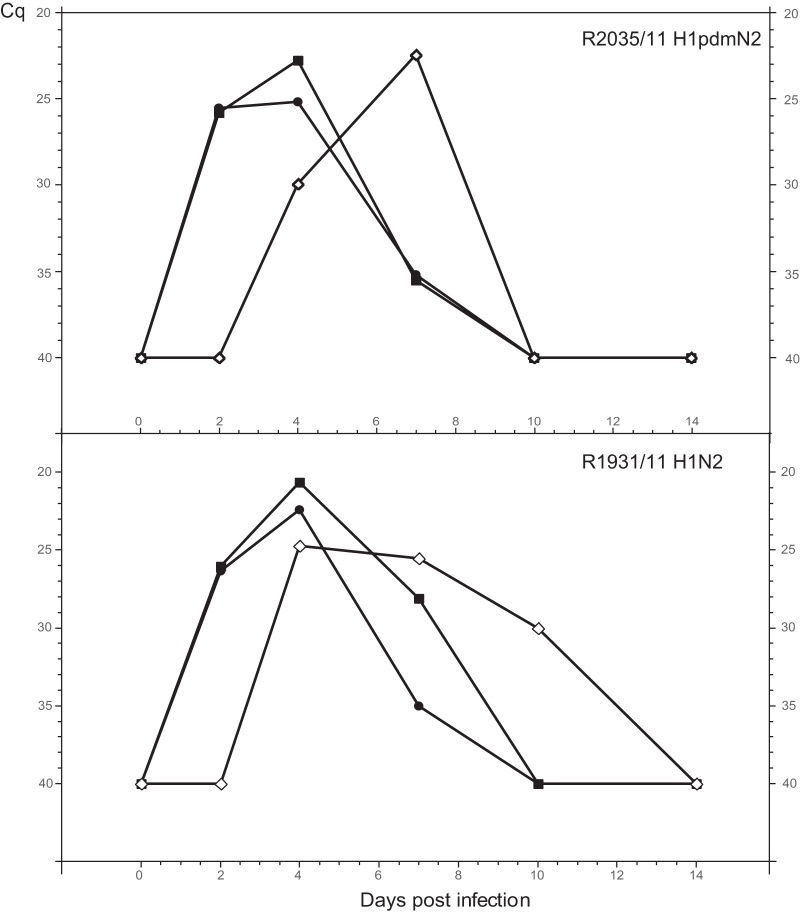
Two recent porcine isolates were selected for experimental infection studies with minipigs. Isolate R2035/11 (H1pdmN2) belonged to the group of reassortants between pandemic H1N1 and porcine HxN2 viruses that have emerged recently. As shown in Fig. 5 (top), intranasal inoculation of this isolate led to a productive infection, with significant viral RNA excretion during the first week postinfection. Virus was also transmitted to an in-contact pig, which showed a similar but shifted course of viral RNA excretion. Mild respiratory symptoms, such as sneezing and clear nasal discharge, were observed for all animals around day 5 of infection. None of the pigs showed rectal temperatures exceeding 39.8°C.
Fig 5.
Experimental infections of minipigs. Shown is the course of infection with reassortant virus R2035/11 (H1pdmN2) or R1931/11 (H1avN2), as reflected by the detection of viral RNA in nasal swabs by M-specific RT-qPCR (Cq values). Filled symbols, animals inoculated intranasally; open diamond, sentinel animal, placed with the inoculated animals on day 1 p.i.
Isolate R1931/11 (H1N2) represented a reassortant between an H1N1 virus of avian origin and a porcine HxN2 virus. This isolate revealed several conspicuous genetic and phylogenetic features, described above, and was also antigenically distinguishable from other H1av viruses (Fig. 4). Figure 5 (bottom) shows a course of nasal viral RNA excretion similar to that described for R2035/11. The infection of the contact pig confirms the presence of productive infection leading to rapid transmission of virus. Clinical symptoms were similar to those of the R2035/11 group but were restricted to day 3 only. Likewise, fever was not noted.
Seroconversion by an NP-specific blocking ELISA was evident from day 7 p.i. on for the R1931 group and from day 14 on also by a homologous HI assay for both groups (data not shown). For unknown reasons, no clear-cut seroconversion (days 7 and 14 p.i.) was evident for the R2035 group when the NP-specific blocking ELISA was used.
DISCUSSION
Porcine IAV infections in swine were frequently detected by virological means in studies A and B in a region of Germany with a high intensity of swine farming. Virus was found to circulate primarily in weaners with respiratory symptoms. This is in line with previous studies on the epidemiology of swine influenza viruses that have emphasized the importance of younger pigs as a motor of IAV infections in swine herds (39, 40). However, the breeding sow population has also been implicated as a reservoir of respiratory pathogens (41), and influenza viruses have been associated with abortion and other fertility disorders in sows (42, 43). In order to investigate such associations, study B was conducted, focusing on large swine herds with reported fertility disorders and with records of influenza vaccination. The data are representative for swine holdings that keep sows but not for pure fattening holdings. Our results revealed that virologically detectable infection of older sows was a rare event (5.8% of samples were positive), yet circulating influenza virus was detected in the weaners of such holdings (32.2% of samples were positive). As such, no direct link between influenza virus infections and reproduction failures in older sows could be verified in this study. However, short febrile episodes caused by limited and short-lived virus replication, such as that observed during infection experiments (Fig. 5), may have passed virologically unnoticed. Still, such episodes may have resulted in hormonal imbalances leading to abortion. Influenza viruses also play a significant role in the porcine respiratory disease complex (44), a difficult-to-define syndrome of multifactorial etiology in which complex and often synergistic interactions of different pathogens, including PRRSV and PCV-2, and environmental factors play a role (45–49). Interestingly, evaluation of the questionnaire in study B suggested that vaccination of sows against PRRSV and vaccination of weaners against PCV-2 had substantial effects in reducing the frequency of virologically detectable influenza virus infections in herds in which the sows had received continuous influenza vaccination. These findings prompt the further study of coinfections or preceding infections with these viruses or other bacterial pathogens, which were not specifically investigated in this study.
The passive surveillance carried out in the frame of study B confirmed year-round viral activity for endemic porcine influenza viruses (Fig. 1). This is in contrast to the seasonal pattern of human influenza in temperate zones. Detection in swine of the human pandemic H1N1 virus of 2009 and its reassortants outside the human influenza season (May to December) indicated independent circulation of these viruses in the swine population under study (Table 1). Another study, which involved five European countries excluding Germany, also reported year-round influenza virus activity in swine herds, with peaks in December and May (49). This is corroborated by serological data collected over a 3-year period in four European countries showing only marginal seasonal variation of seroprevalences (50). All-year influenza virus activity in swine herds entails a continuing risk of transmission of porcine influenza viruses from swine to humans. It is worth noting that three infections of humans in Germany with influenza viruses that occurred outside the usual influenza season in 2010 and 2011 were in fact caused by endemic porcine influenza viruses (51).
Characterization of porcine IAV revealed the presence of the three standard European subtypes H1N1, H1N2, and H3N2. Nasal swab samples obtained from acutely infected pigs were preferable to lung tissue samples from terminally diseased swine for subtype-specific characterization (77.9% versus 30.2% of fully subtyped samples were positive [Table 1]) but not for generic detection of swine influenza viruses (21.4% versus 24.4%). This was largely due to the fact that as many as 10 nasal swab samples from a herd, in contrast to 1 or 2 tissue samples, were available for the selection of samples with higher contents of viral RNA for subtyping. The quality of RNA from swabs versus lung tissues, however, did not cause this difference in successful subtyping. Viruses of the avian influenza virus-derived H1N1 lineage made up almost two-thirds of the fully subtyped samples. H1N2 and H3N2 viruses were detected at much lower prevalences (17.6 and 11.1%, respectively). The dominance of H1av-like viruses is in line with earlier reports from Germany (6, 52) and other European countries (49). Viruses of the human pandemic H1N1/2009 virus have been repeatedly introduced from the human population into swine herds in Germany since 2009 (9). Considering data that demonstrated high susceptibility of swine to this lineage and ease of transmission between pigs (18, 40), it seems odd that only two herds with H1N1pdm infections were found during the surveillance period (Table 1). No immunization against this lineage had been carried out during the study period. However, reassortants of H1pdm viruses with porcine HxN2 viruses emerged in this region in May 2011 (10) and continued to circulate until the end of the study period, when 11 herds with H1pdmN2 infections were found. It is tempting to speculate that H1pdmN2 viruses have evolved a selective advantage over the H1N1pdm lineage and are therefore spreading preferentially. In countries where such reassortants have not emerged, e.g., the United Kingdom, H1N1pdm viruses have obviously gained more ground (40). The hemagglutinin of most isolates of the H1pdmN2 reassortant viruses clustered separately from 2009 H1N1pdm hemagglutinin in a new clade. Both the HA and the NA of these viruses are characterized by a large set of amino acid substitutions that separate them from their ancestral sequences. Since an optimal balance between the functions of the counteracting HA and NA glycoproteins is required for fitness of the virus, it may be speculated that the substitutions recorded represent the current result of mutual adaptations to the new partner protein.
The phylogenetic analyses of HA1 and NA sequences provided further insight into the diversity of cocirculating porcine influenza viruses: recent H1N2 and H3N2 HA1 sequences formed monophyletic clades that were temporally (H3N2) or geographically (H1N2) separated from their nearest neighbors in the trees (Fig. 2d; see also Fig. S2 in the supplemental material). The H1N2 viruses in particular seem to have undergone separate evolutions in different European countries. This seems to point toward transmission chains that do not cross political borders in the European Common Market. In this respect, studies on networks in the swine trade in several European countries point out that the vast majority of trading actions are compartmentalized and restricted to a few spatially clustered communities, with rare contacts outside these trading pools (53, 54). This might explain to some extent the geographic restriction of porcine influenza virus evolution, although it is not clear why this should affect predominantly viruses of subtype H1N2.
Complex branching patterns were evident for the HA1 fragment of H1 of avian origin. At least four sublineages were discernible on the basis of nucleotide sequences, and some could be distinguished by unique amino acid substitution patterns as well. Similar diverging trends have also been noticed for the classical porcine H1N1 virus in North America (55). In the current study, one group of related viruses (R369/09, R848/11, R1931/11, R3310/11) was located on unusually long horizontal branches and showed a particular accumulation of amino acid substitutions. These also affected the pattern of potential N-linked glycosylation sites, which is known to have an impact on antigenicity and immunogenicity (56, 57). N1 sequences of porcine influenza viruses from Germany fell into three lineages, all of which were derived from the same avian precursor. The origin of N2 sequences was assigned to four lineages. H1pdmN2 viruses had derived their N2 from three of these lineages (10). One lineage was derived from precursor viruses prevalent in the human population during the mid-1990s. No other porcine viruses carrying a neuraminidase of this lineage have been described so far. Since this lineage also seems extinct in the human population, the reservoir of this neuraminidase remains obscure.
Nine reassortment patterns involving surface glycoprotein-encoding segments of the endemic H1N1av, H1N2, and H3N2 Eurasian porcine subtypes were detected in this study over a surveillance period of 19 months. This is a rate of 10.2% given a set of 88 fully subtyped viruses. Since fewer NA than HA sequences were generated in this study, this figure may still underrepresent the reassorted viruses actually in circulation. Yet the proportion of reassorted viruses observed here is higher, and their diversity wider, than those recently estimated for Eurasian swine influenza viruses (58) and recently shown in field data by Kyriakis et al. (49), who reported a reassortment rate of 3% over a 3-year surveillance period in samples from five European countries excluding Germany. The comparatively frequent incidence of HA/NA reassortments is also reflected by the fact that in five cases, infections with two different subtypes were detected at the same time in the same herd but in different animals (Table 1).
In contrast to the variability of HA and NA and their frequent reassortments, the polymerase complex and the remaining “internal” gene segments of recent porcine IAVs from Germany seem to be comparatively stable with respect to their sequences and their gene constellations (Table 2; see also Fig. S2 in the supplemental material). Three genogroups were distinguished in this study based on the limited number of 18 viruses that were fully sequenced. The only segment that showed reassortment, apart from HA and NA, was NS, in two cases. This finding is in line with recent analyses of Eurasian swine influenza virus sequences indicating that the current set of “internal” gene segments of the avian influenza virus-derived Eurasian lineage (Table 2, genogroup 1), and here especially the polymerase complex, seems to form a stable constellation similar to the TRIG cassette of North American porcine viruses (58). However, in contrast to the situation in the United States, where frequent reassortants of “internal segments” between the pandemic H1N1pdm virus and American endemic porcine viruses have been found (59), no such reassortants were detected in the present study. Genogroup 3, which represents the standard set of segments of H1pdm viruses, was also retained in the H1pdmN2 reassortant viruses analyzed here. Genogroup 2 (R369/09 and R1931/11), however, appears to typify another gene constellation, which has not been described before. These viruses, together with R3310/11 and R848/11, form a loose group of isolates with an H1av HA and either an N1av or an N2hu NA. In addition, in one of these viruses (R1931/11), aa 147 in the HA protein was deleted and aberrant patterns of potential NGSs were expressed. The NS1 gene segment of this isolate harbored a variant PDV domain (RPKV) and could not be typed with the FluGenome genotyping tool; its PB1-F2 segment expressed the rare N66S substitution.
The different character of isolate R1931/11 is also reflected in its distinct antigenic properties, which were demonstrated by HI using a panel of polyclonal postinfection sera from swine and ferrets. The normalized cross-reactive HI titer panel, depicted as a heat map and a corresponding dendrogram in Fig. 4, allows the classification of the standard subtype H1 viruses into avian influenza virus-derived H1av, pandemic H1pdm, and H1N2 viruses. While the H1N2 isolates formed a homogenous block, lineages were discernible among the H1av and H1pdm viruses. In the H1pdm lineage, H1pdmN2 reassortants were distinguishable from “standard” H1pdm viruses circulating in swine and humans (R26/11) in Germany. Within the H1av lineage, two antigenically distinct groups were evident. R1931/11 is associated with one of these H1av groups but forms a separate branch. The deletion ΔK147 (according to H1 numbering), detected solely in this isolate, exactly matches position K134 (according to H3 numbering) in human seasonal H1 viruses (Table 3). Deletion of K134 has been shown to cause significant antigenic drift in those strains (60). The segregation of R1931/11 suggested on the basis of the HI antigenic properties was fully reflected by the phylogenetic analysis of the deduced HA1 amino acid sequences of these viruses (Fig. 4, left dendrogram). The epidemiological relevance of diverging antigenic trends in H1av and H1pdm viruses remains to be analyzed. This analysis would include vaccination-challenge experiments to explore the cross-protective versus neutralization escape tendencies of the emerging lineages.
Table 3.
Deletion of lysine residue K147 (H1 numbering) in the HA1 fragment of R1931/11 (H1avN2) matches deletion K134 (H3 numbering) in human seasonal H1 viruses known to have caused antigenic drifta
| Virus | Sequence alignmentb |
|---|---|
| A/Beijing/262/95 (H1N1) | 126-S W P N H T V T - G V T A S C |
| A/Shenzhen/227/95 (H1N1) | 126-S W P N H T V T K G V T A S C |
| A/Swine/Germany/R1931/11 (H1avN2) | 139-S W P D H K T T - G T T G S C |
| A/Swine/Germany/R655/12 (H1N1av) | 139-S W P N H E T T K G S T V A C |
See reference 60.
Boldface lettering indicates the deletion site.
Two viruses from lineages with conspicuous genetic, phylogenetic, and antigenic alterations from the standard European subtypes were selected for infection experiments with minipigs. Both the H1pdmN2 isolate R2035/11 and the aberrant H1avN2 reassortant R1931/11 replicated productively in pigs following intranasal inoculation and were readily transmitted to an in-contact sentinel. These findings indicate the potential of these viruses for further spread in the swine population. The very mild clinical course observed should not be overinterpreted, since minipigs from a closed breeding nucleus may react differently from pigs of standard lineages and may show milder clinical symptoms. For example, minipigs did not show clinical symptoms when challenged with the California/04 H1N1pdm isolate (61), which, in contrast, produced moderate symptoms similar to those of endemic porcine influenza in standard pigs (62, 63). Moreover, the minipigs were 13 weeks old when inoculated, and their increased age, compared to that of younger weanling pigs, may explain the mild clinical signs.
In summary, we have shown frequent, widespread, and year-round circulation of at least four stable lineages (H1N1, H1N2, H3N2, H1pdmN2) of porcine influenza viruses and the focal presence of further reassortants between these lineages in a region of northwestern Germany that has 55% of the country's pig population. Trends toward ongoing diversification at both the phylogenetic and antigenic levels were demonstrated for the H1N1av lineage and its reassortants, which still dominated the epidemiological situation. In addition, continuing diverging evolution was also noticed for the recently emerging H1pdmN2 lineage. These viruses carry seven segments of the human pandemic H1N1 2009 virus, including a markedly antigenically altered hemagglutinin and different variants of pig- or human-derived subtype N2 neuraminidase. The zoonotic potential of this lineage remains to be determined. Tight control of influenza virus infections by improved herd management, highly efficacious vaccines, and effective herd vaccination regimens is highly desirable for the health and productive potential of swine herds. Regular influenza virus surveillance of swine populations is a pivotal prerequisite of this goal. In line with the One Health concept, reduced circulation of influenza viruses in pigs also means lowered risks of the formation of new reassortant viruses with unknown zoonotic potential and of their transmission to humans.
Supplementary Material
ACKNOWLEDGMENTS
This study depended on the cooperation and openness of attending veterinarians and swine holders who sent in samples and supplied accompanying epidemiological information. We are grateful for the excellent technical support of Sven Sander, Katja Hartwig, Anne Carnitz, Martina Abs, and Kathrin Steffen. We also acknowledge the authors and the laboratories that originated and submitted the sequences from GISAID's EpiFlu database on which this research is based. The list is given in Table S4 in the supplemental material.
Financial support was provided by a grant from vaxxinova GmbH, Cuxhaven, Germany, to the FLI and to the University of Veterinary Medicine, Hannover, Germany. The FLI was further funded by a grant from the Federal Ministry for Food, Agriculture and Consumer Protection (FSI-2-3.2), the European Union FP7 project European Management Platform for Emerging and Re-emerging Infectious Disease Entities (EMPERIE; grant 223498), and European Union FP7 project ESNIP3.
Footnotes
Published ahead of print 3 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00381-13.
REFERENCES
- 1.Olsen CW, Brown IH, Easterday BC, van Reeth K. 2006. Swine influenza, p 469–482 In Straw BE, Zimmerman JJD', Allaire S, Taylor DJ. (ed), Diseases of swine, 9th ed. Blackwell Publishing, Oxford, United Kingdom [Google Scholar]
- 2.Thacker E, Janke B. 2008. Swine influenza virus: zoonotic potential and vaccination strategies for the control of avian and swine influenzas. J. Infect. Dis. 197(Suppl 1):S19–S24 [DOI] [PubMed] [Google Scholar]
- 3.Torremorell M, Allerson M, Corzo C, Diaz A, Gramer M. 9 January 2012. Transmission of influenza A virus in pigs. Transbound. Emerg. Dis. [Epub ahead of print.] 10.1111/j.1865-1682.2011.01300.x [DOI] [PubMed] [Google Scholar]
- 4.Irvine RM, Brown IH. 2009. Novel H1N1 influenza in people: global spread from an animal source? Vet. Rec. 164:577–578 [DOI] [PubMed] [Google Scholar]
- 5.Brown IH. 2013. History and epidemiology of swine influenza in Europe. Curr. Top. Microbiol. Immunol. 370:133–146 [DOI] [PubMed] [Google Scholar]
- 6.Van Reeth K, Brown IH, Dürrwald R, Foni E, Labarque G, Lenihan P, Maldonado J, Markowska-Daniel I, Pensaert M, Pospisil Z, Koch G. 2008. Seroprevalence of H1N1, H3N2 and H1N2 influenza viruses in pigs in seven European countries in 2002–2003. Influenza Other Respi. Viruses 2:99–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuntz-Simon G, Madec F. 2009. Genetic and antigenic evolution of swine influenza viruses in Europe and evaluation of their zoonotic potential. Zoonoses Public Health 56:310–325 [DOI] [PubMed] [Google Scholar]
- 8.Brockwell-Staats C, Webster RG, Webby RJ. 2009. Diversity of influenza viruses in swine and the emergence of a novel human pandemic influenza A (H1N1). Influenza Other Respi. Viruses 3:207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Starick E, Lange E, Fereidouni S, Bunzenthal C, Höveler C, Kuczka A, grosse Beilage E, Hamann HP. 2011. Re-assorted pandemic (H1N1) 2009 influenza A virus discovered from pigs in Germany. J. Gen. Virol. 92:1184–1188 [DOI] [PubMed] [Google Scholar]
- 10.Starick E, Lange E, Grund C, grosse Beilage E, Döhring S, Maas A, Noé T, Beer M, Harder TC. 2012. Reassortants of pandemic influenza A virus H1N1/2009 and endemic porcine HxN2 viruses emerge in swine populations in Germany. J. Gen. Virol. 93:1658–1663 [DOI] [PubMed] [Google Scholar]
- 11.Kitikoon P, Vincent AL, Jones KR, Nilubol D, Yu S, Janke BH, Thacker BJ, Thacker EL. 2009. Vaccine efficacy and immune response to swine influenza virus challenge in pigs infected with porcine reproductive and respiratory syndrome virus at the time of SIV vaccination. Vet. Microbiol. 139:235–244 [DOI] [PubMed] [Google Scholar]
- 12.Loeffen WL, Heinen PP, Bianchi AT, Hunneman WA, Verheijden JH. 2003. Effect of maternally derived antibodies on the clinical signs and immune response in pigs after primary and secondary infection with an influenza H1N1 virus. Vet. Immunol. Immunopathol. 92:23–35 [DOI] [PubMed] [Google Scholar]
- 13.Vijaykrishna D, Smith GJ, Pybus OG, Zhu H, Bhatt S, Poon LL, Riley S, Bahl J, Ma SK, Cheung CL, Perera RA, Chen H, Shortridge KF, Webby RJ, Webster RG, Guan Y, Peiris JS. 2011. Long-term evolution and transmission dynamics of swine influenza A virus. Nature 473:519–522 [DOI] [PubMed] [Google Scholar]
- 14.Fereidouni SR, Harder TC, Gaidet N, Ziller M, Hoffmann B, Hammoumi S, Globig A, Starick E. 2012. Saving resources: avian influenza surveillance using pooled swab samples and reduced reaction volumes in real-time RT-PCR. J. Virol. Methods 186:119–125 [DOI] [PubMed] [Google Scholar]
- 15.Hoffmann B, Depner K, Schirrmeier H, Beer M. 2006. A universal heterologous internal control system for duplex real-time RT-PCR assays used in a detection system for pestiviruses. J. Virol. Methods 136:200–209 [DOI] [PubMed] [Google Scholar]
- 16.Gall A, Hoffmann B, Harder T, Grund C, Beer M. 2008. Universal primer set for amplification and sequencing of HA0 cleavage sites of all influenza A viruses. J. Clin. Microbiol. 46:2561–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gall A, Hoffmann B, Harder T, Grund C, Ehricht R, Beer M. 2009. Rapid and highly sensitive neuraminidase subtyping of avian influenza viruses by use of a diagnostic DNA microarray. J. Clin. Microbiol. 47:2985–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lange E, Kalthoff D, Blohm U, Teifke JP, Breithaupt A, Maresch C, Starick E, Fereidouni S, Hoffmann B, Mettenleiter TC, Beer M, Vahlenkamp TW. 2009. Pathogenesis and transmission of the novel swine-origin influenza virus A/H1N1 after experimental infection of pigs. J. Gen. Virol. 90:2119–2123 [DOI] [PubMed] [Google Scholar]
- 19.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275–2289 [DOI] [PubMed] [Google Scholar]
- 20.Li OT, Barr I, Leung CY, Chen H, Guan Y, Peiris JS, Poon LL. 2007. Reliable universal RT-PCR assays for studying influenza polymerase subunit gene sequences from all 16 haemagglutinin subtypes. J. Virol. Methods 142:218–222 [DOI] [PubMed] [Google Scholar]
- 21.Katoh K, Asimenos G, Toh H. 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 537:39–64 [DOI] [PubMed] [Google Scholar]
- 22.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253–1256 [DOI] [PubMed] [Google Scholar]
- 24.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 25.Smith GJD, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JSM, Guan Y, Rambaut A. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122–1125 [DOI] [PubMed] [Google Scholar]
- 26.Seo J, Gordish-Dressman H, Hoffman EP. 2006. An interactive power analysis tool for microarray hypothesis testing and generation. Bioinformatics 22:808–814 [DOI] [PubMed] [Google Scholar]
- 27.Lai AC, Wu WL, Lau SY, Guan Y, Chen H. 2012. Two-dimensional antigenic dendrogram and phylogenetic tree of avian influenza virus H5N1. FEMS Immunol. Med. Microbiol. 64:205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitley RJ, Boucher CA, Lina B, Nguyen-Van-Tam J, Osterhaus A, Schutten M, Monto AS. 2013. Global assessment of resistance to neuraminidase inhibitors, 2008–2011: the Influenza Resistance Information Study (IRIS). Clin. Infect. Dis. 56:1197–1205 [DOI] [PubMed] [Google Scholar]
- 29.Lu G, Rowley T, Garten R, Donis RO. 2007. FluGenome: a web tool for genotyping influenza A virus. Nucleic Acids Res. 35(Web Server issue):W275–W279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunham EJ, Dugan VG, Kaser EK, Perkins SE, Brown IH, Holmes EC, Taubenberger JK. 2009. Different evolutionary trajectories of European avian H1N1-like and classical swine H1N1 influenza A viruses. J. Virol. 83:5485–5494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krumbholz A, Philipps A, Oehring H, Schwarzer K, Eitner A, Wutzler P, Zell R. 2011. Current knowledge on PB1-F2 of influenza A viruses. Med. Microbiol. Immunol. 200:69–75 [DOI] [PubMed] [Google Scholar]
- 32.Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. 2007. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 3:1414–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi M, Jagger BW, Wise HM, Digard P, Holmes EC, Taubenberger JK. 2012. Evolutionary conservation of the PA-X open reading frame in segment 3 of influenza A virus. J. Virol. 86:12411–12413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moreno A, Chiapponi C, Boniotti MB, Sozzi E, Foni E, Barbieri I, Zanoni MG, Faccini S, Lelli D, Cordioli P. 2012. Genomic characterization of H1N2 swine influenza viruses in Italy. Vet. Microbiol. 156:265–276 [DOI] [PubMed] [Google Scholar]
- 36.Obenauer JC, Denson J, Mehta PK, Su X, Mukatira S, Finkelstein DB, Xu X, Wang J, Ma J, Fan Y, Rakestraw KM, Webster RG, Hoffmann E, Kraus S, Zheng J, Zhang Z, Naeve CW. 2006. Large-scale sequence analysis of avian influenza isolates. Science 311:1576–1580 [DOI] [PubMed] [Google Scholar]
- 37.Krumbholz A, Schmidtke M, Bergmann S, Motzke S, Bauer K, Stech J, Dürrwald R, Wutzler P, Zell R. 2009. High prevalence of amantadine resistance among circulating European porcine influenza A viruses. J. Gen. Virol. 90:900–908 [DOI] [PubMed] [Google Scholar]
- 38.Wiley DC, Wilson IA, Shekel JJ. 1981. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289:373–378 [DOI] [PubMed] [Google Scholar]
- 39.Takemae N, Parchariyanon S, Ruttanapumma R, Hiromoto Y, Hayashi T, Uchida Y, Saito T. 2011. Swine influenza virus infection in different age groups of pigs in farrow-to-finish farms in Thailand. Virol. J. 8:537. 10.1186/1743-422X-8-537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williamson SM, Tucker AW, McCrone IS, Bidewell CA, Brons N, Habernoll H, Essen SC, Brown IH, COSI. Wood JL. 2012. Descriptive clinical and epidemiological characteristics of influenza A H1N1 2009 virus infections in pigs in England. Vet. Rec. 171:271. 10.1136/vr.100673 [DOI] [PubMed] [Google Scholar]
- 41.Fablet C, Marois C, Kuntz-Simon G, Rose N, Dorenlor V, Eono F, Eveno E, Jolly JP, Le Devendec L, Tocqueville V, Quéguiner S, Gorin S, Kobisch M, Madec F. 2011. Longitudinal study of respiratory infection patterns of breeding sows in five farrow-to-finish herds. Vet. Microbiol. 147:329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madec F, Kaiser C, Gourreau JM, Martinat-Botte F. 1989. Pathologic consequences of a severe influenza outbreak (swine virus A/H1N1) under natural conditions in the non-immune sow at the beginning of pregnancy. Comp. Immunol. Microbiol. Infect. Dis. 12:17–27 (In French.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wesley RD. 2004. Exposure of sero-positive gilts to swine influenza virus may cause a few stillbirths per litter. Can. J. Vet. Res. 68:215–217 [PMC free article] [PubMed] [Google Scholar]
- 44.Opriessnig T, Giménez-Lirola LG, Halbur PG. 2011. Polymicrobial respiratory disease in pigs. Anim. Health Res. Rev. 12:133–148 [DOI] [PubMed] [Google Scholar]
- 45.Ellis J, Clark E, Haines D, West K, Krakowka S, Kennedy S, Allan GM. 2004. Porcine circovirus-2 and concurrent infections in the field. Vet. Microbiol. 98:159–163 [DOI] [PubMed] [Google Scholar]
- 46.Jung K, Renukaradhya GJ, Alekseev KP, Fang Y, Tang Y, Saif LJ. 2009. Porcine reproductive and respiratory syndrome virus modifies innate immunity and alters disease outcome in pigs subsequently infected with porcine respiratory coronavirus: implications for respiratory viral co-infections. J. Gen. Virol. 90:2713–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wellenberg GJ, Bouwkamp FT, Wolf PJ, Swart WA, Mombarg MJ, de Gee AL. 2010. A study on the severity and relevance of porcine circovirus type 2 infections in Dutch fattening pigs with respiratory diseases. Vet. Microbiol. 142:217–224 [DOI] [PubMed] [Google Scholar]
- 48.Deblanc C, Robert F, Pinard T, Gorin S, Quéguiner S, Gautier-Bouchardon AV, Ferré S, Garraud JM, Cariolet R, Brack M, Simon G. 29 November 2012, posting date Pre-infection of pigs with Mycoplasma hyopneumoniae induces oxidative stress that influences outcomes of a subsequent infection with a swine influenza virus of H1N1 subtype. Vet. Microbiol. 10.1016/j.vetmic.2012.11.028 [DOI] [PubMed] [Google Scholar]
- 49.Kyriakis CS, Brown IH, Foni E, Kuntz-Simon G, Maldonado L, Madec F, Essen SC, Chiapponi C, Van Reeth K. 2011. Virological surveillance and preliminary antigenic characterization of influenza viruses in pigs in five European countries from 2006 to 2008. Zoonoses Public Health 58:93–101 [DOI] [PubMed] [Google Scholar]
- 50.Kyriakis CS, Rose N, Foni E, Maldonado J, Loeffen WLA, Madec F, Simon G, Van Reeth K. 14 November 2012, posting date Influenza A virus infection dynamics in swine farms in Belgium, France, Italy and Spain, 2006-2008 Vet. Microbiol. 10.1016/j.vetmic.2012.11.014 [DOI] [PubMed] [Google Scholar]
- 51.Buda S, Haas W, Baillot A, Beyrer K, Monazahian M, Pulz M, Benzler J, Harder TC, Schweiger B. 2011. Human infections with porcine influenza viruses. Epidemiol. Bull. RKI 39:357–359 (In German.) [Google Scholar]
- 52.Lang C, Palzer A, Dürrwald R, Selbitz HJ, Ritzmann M. 2010. Analysis of the situation of swine influenza in the districts Upper Bavaria, Swabia, Freiburg and Tübingen based on antibody profiles of piglets from Bavarian breeding farms. Berl. Munch. Tierarztl. Wochenschr. 123:385–391 (In German.) [PubMed] [Google Scholar]
- 53.Lentz HH, Konschake M, Teske K, Kasper M, Rother B, Carmanns R, Petersen B, Conraths FJ, Selhorst T. 2011. Trade communities and their spatial patterns in the German pork production network. Prev. Vet. Med. 98:176–181 [DOI] [PubMed] [Google Scholar]
- 54.Rautureau S, Dufour B, Durand B. 2012. Structural vulnerability of the French swine industry trade network to the spread of infectious diseases. Animal 6:1152–1162 [DOI] [PubMed] [Google Scholar]
- 55.Nfon CK, Berhane Y, Hisanaga T, Zhang S, Handel K, Kehler H, Labrecque O, Lewis NS, Vincent AL, Copps J, Alexandersen S, Pasick J. 2011. Characterization of H1N1 swine influenza viruses circulating in Canadian pigs in 2009. J. Virol. 85:8667–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schulze IT. 1997. Effects of glycosylation on the properties and functions of influenza virus hemagglutinin. J. Infect. Dis. 176(Suppl 1):S24–S28 [DOI] [PubMed] [Google Scholar]
- 57.Vigerust DJ, Shepard VL. 2007. Virus glycosylation: role in virulence and immune interaction. Trends Microbiol. 15:211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lycett SJ, Bailie G, Coulter E, Bhatt S, Kellam P, McCauley JW, Wood JL, Brown IH, Pybus OG, Leigh Brown AJ. 2012. Estimating reassortment rates in co-circulating Eurasian swine influenza viruses. J. Gen. Virol. 93:2326–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ducatez MF, Hause B, Stigger-Rosser E, Darnell D, Corzo C, Juleen K, Simonson R, Brockwell-Staats C, Rubrum A, Wang D, Webb A, Crumpton JC, Lowe J, Gramer M, Webby RJ. 2011. Multiple reassortment between pandemic (H1N1) 2009 and endemic influenza viruses in pigs, United States. Emerg. Infect. Dis. 17:1624–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McDonald NJ, Smith CB, Cox NJ. 2007. Antigenic drift in the evolution of H1N1 influenza A viruses resulting from deletion of a single amino acid in the haemagglutinin gene. J. Gen. Virol. 88:3209–3213 [DOI] [PubMed] [Google Scholar]
- 61.Itoh Y, Shinya K, Kiso M, Watanabe T, Sakoda Y, Hatta M, Muramoto Y, Tamura D, Sakai-Tagawa Y, Noda T, Sakabe S, Imai M, Hatta Y, Watanabe S, Li C, Yamada S, Fujii K, Murakami S, Imai H, Kakugawa S, Ito M, Takano R, Iwatsuki-Horimoto K, Shimojima M, Horimoto T, Goto H, Takahashi K, Makino A, Ishigaki H, Nakayama M, Okamatsu M, Takahashi K, Warshauer D, Shult PA, Saito R, Suzuki H, Furuta Y, Yamashita M, Mitamura K, Nakano K, Nakamura M, Brockman-Schneider R, Mitamura H, Yamazaki M, Sugaya N, Suresh M, Ozawa M, Neumann G, Gern J, Kida H, Ogasawara K, Kawaoka Y. 2009. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460:1021–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brookes SM, Núñez A, Choudhury B, Matrosovich M, Essen SC, Clifford D, Slomka MJ, Kuntz-Simon G, Garcon F, Nash B, Hanna A, Heegaard PM, Quéguiner S, Chiapponi C, Bublot M, Garcia JM, Gardner R, Foni E, Loeffen W, Larsen L, Van Reeth K, Banks J, Irvine RM, Brown IH. 2010. Replication, pathogenesis and transmission of pandemic (H1N1) 2009 virus in non-immune pigs. PLoS One 5:e9068. 10.1371/journal.pone.0009068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vincent AL, Lager KM, Faaberg KS, Harland M, Zanella EL, Ciacci-Zanella JR, Kehrli ME, Jr, Janke BH, Klimov A. 2010. Experimental inoculation of pigs with pandemic H1N1 2009 virus and HI cross-reactivity with contemporary swine influenza virus antisera. Influenza Other Respi. Viruses 4:53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.