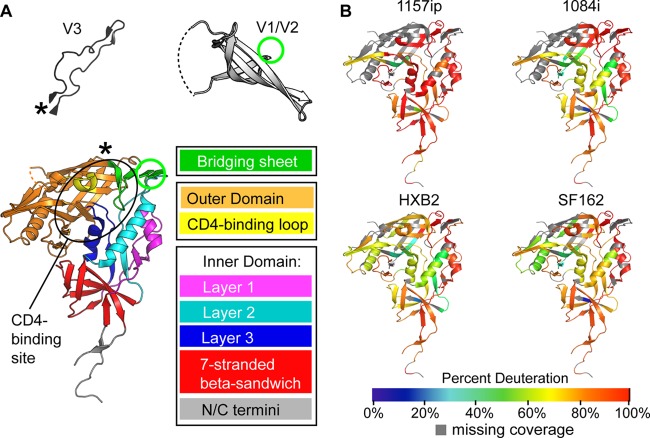
Fig 2.
Summary of isolate-specific differences in unliganded gp120 conformational dynamics. (A) The organization of monomeric gp120 into inner, outer, and bridging sheet domains is indicated by colors on the gp120 core with N- and C-terminal extensions (PDB ID 3JWD). The inner domain is further divided into mobile layers 1, 2, and 3 (magenta, cyan, and blue, respectively) that protrude from the 7-stranded β-sandwich (red) (22). Variable loops V1/V2 and V3 (from PDB 3U4E and PDB 2B4C, respectively) are included for reference. The positions of these elements in the context of the gp120 core are indicated by an asterisk for V3 and a green circle for V1/V2. The approximate location of the CD4 binding site is indicated with a black oval. The structures shown here and throughout the text reflect that of gp120 in a receptor-bound conformation, because no structure is currently available for full-length, unliganded gp120. These structures may not necessarily reflect the unliganded structure of full-length gp120 in solution. (B) Heat maps of HDX-MS data for unliganded gp120 reveal qualitative isolate-specific differences in structural dynamics. Colors mapped onto the gp120 core (PDB ID 3JWD) indicate the percent deuteration of peptides after 1 min of incubation in a deuterated buffer; warm colors correspond to high levels of deuterium uptake (dynamic regions), and cool colors correspond to low levels of deuteration (ordered or “protected” regions). Regions where peptide information is missing are indicated in gray.