Fig 1.
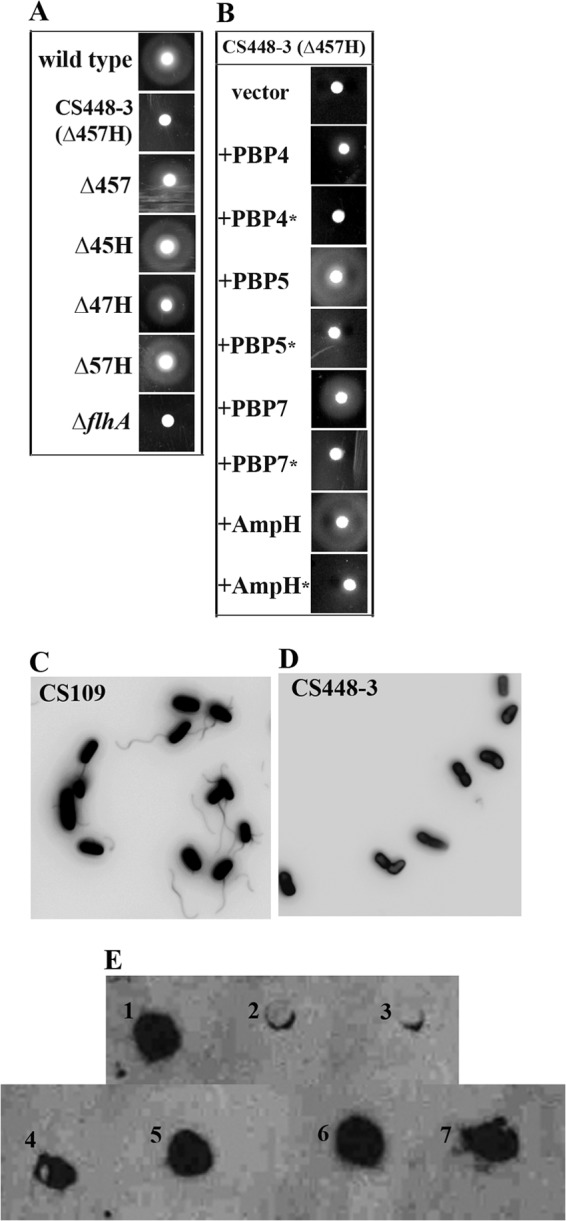
Loss of four specific PBPs inhibits migration by inhibiting flagellum production. The migration of each strain was assayed over 10 h on migration agar. (A) E. coli strain CS448-3, lacking PBPs 4, 5, and 7 and AmpH (Δ457H, row 2), is nonmotile after 10 h. Strains lacking any three of these PBPs remained motile: Δ457 (CS315-1), Δ45H (CS326-3), Δ47H (CS317-3), and Δ57H (CS345-3). A strain unable to make flagella, an ΔflhA mutant strain (SKCS49-1), served as a negative control. (B) Complementation with any one wild-type PBP gene restores the ability to migrate to E. coli CS448-3. CS448-3 was transformed with the following plasmids (the wild-type PBP that was expressed is noted in parentheses): pLPKC403 (PBP4, row 2), pLP515 (PBP5, row 4), pLPKC704 (PBP7, row 6), and pLCM1 (AmpH, row 8). No complementation occurred when PBPs with mutated active sites were expressed: pLP405 (PBP4*, row 3), pLP514 (PBP5*, row 5), pLP706 (PBP7*, row 7), and pMEL1 (AmpH*, row 9). CS448-3 transformed with plasmid vectors was nonmotile: CS448-3 pLP18 (top row) and CS448-3 pLP8 (data not shown). The parental strain containing plasmid vectors was motile (data not shown). (C and D) PBP loss inhibits flagellum production. E. coli CS109 (C) and CS448-3 (D) were stained with Alexa Fluor 488. Wild-type cells displayed at least one flagellum per cell (C), whereas CS448-3 lacked flagella (D). (E) Whole cells were spotted onto a PVDF membrane prewetted in methanol, and the FliC flagellar protein was detected by Western blotting. E1, CS109; E2, CS448-3; E3, CS109ΔflhA (SKCS49-1); E4, CS109ΔrcsB (KE8); E5, CS448-3ΔrcsB (KE9); E6, CS109/pFlhDC; E7, CS448-3/pFlhDC.
